Graphical abstract
Keywords: Long noncoding RNA, lncRNA, Endogenous, Dark regions, Unannotated, Human genome, Novel
Abstract
>98% of the human genome is composed of noncoding regions and >93% of these noncoding regions are actively transcribed, suggesting their criticality in the human genome. Yet <1% of these regions have been functionally characterized, leaving most of the human genomes in the dark. Here, this study processes petabyte level data and systematically decodes endogenous lncRNAs located in unannotated regions of the human genome and deciphers a distinctive functional regime of lncRNAs hidden in massive RNAseq data. LncRNAs divergently distribute across chromosomes, independent of protein-coding regions. Their transcriptions rarely initiate on promoters through polymerase II, but rather partially on enhancers. Yet conventional enhancer markers (e.g. H3K4me1) only account for a small proportion of lncRNA transcriptions, suggesting alternatively unknown mechanisms initiating the majority of lncRNAs. Furthermore, lncRNA-self regulation also notably contributes to lncRNA activation. LncRNAs regulate broad bioprocesses, including transcription and RNA processing, cell cycle, respiration, response to stress, chromatin organization, post-translational modification, and development. Therefore, lncRNAs functionally govern their own regime distinctive from protein coding genes. This finding establishes a clear framework to comprehend human genome-wide lncRNA-lncRNA and lncRNA-protein coding gene regulations.
Key points.
-
1)
lncRNA genome locations are independent from protein-coding regions.
-
2)
lncRNAs have their own initiation system. lncRNA transcriptions rarely initiate on promoters through polymerase II, but partially on enhancers. Conventional enhancer activators (e.g. H3K4me1) only account for a small proportion of lncRNA transcriptions.
-
3)
lncRNAs are primarily regulated by lncRNAs, instead of proteins, contrary to existing notion.
1. Introduction
Human cells consume enormous energy to transcribe over 93% of their genome [1], which consists of 98% noncoding regions [2]. These noncoding regions were once thought as desert, but increasing evidence demonstrates that they are crucial in human biology [3], [4], [5]. Long noncoding RNAs (lncRNAs, usually >200 bp) [6] predominate the transcripts from noncoding regions [7]. Understanding the abundant lncRNA functions helps appreciate the fundamental functions of the human genome.
The general strategy for characterizing protein-coding mRNAs has been conventionally applied to understand lncRNAs [6], [8]. For example, lncRNA identification has been derived from the concept of protein identification, such as promoter, start codon, poly-A tail and RNA polymerase II (Pol II), and DNA conservation [6], [8]. Combining mRNA concept and sequencing approaches, the GENCODE project V35 [7] has collected 16,899 lncRNAs, in which long intergenic noncoding RNAs (lincRNAs) and antisense RNAs have been merged into a lncRNA category. The FANTOM project has also identified 19,175 lncRNAs from 5′s strategy capturing 5′ mRNA caps [9]. However, these current experimental approaches are biased to experimental conditions like specific cell types and thus they have only identified a limited number of lncRNAs. Bioinformatics and computational tools can help to identify lncRNAs [10], [11], but their development has been slow to systematically identify novel lncRNAs in the human genome, leaving most genome regions in the dark.
On the other hand, most lncRNAs identified to date in humans are tissue-specific [12]. However, a certain number of lncRNAs are evolutionary or functionally conserved. For example, zebrafish carry conserved lncRNAs crucial for embryonic development and these lncRNAs are functionally conserved across species [6]. Mouse genome contains >1,000 lncRNAs with substantial evolutionary conservation (>95%) cross-mammalian [13]. The human genome displays>90% of conserved synteny in the corresponding regions with the mouse genome [14]. Over 71% of human genes possess zebrafish orthologues [15], [16]. In addition, the number of identified lncRNA transcripts increases from zebrafish, mouse to human respectively with 4,852, 131,697 to 172,216 [17]. We hypothesize that the human genome holds a large number of lncRNAs endogenous across all cell-types and tissues and conditions.
This study employed our new software FINET [18] to systematically identify the unannotated lncRNAs (ulncRNAs) endogenous in dark regions of human genome via exhaustively searching a ulncRNA regulatory network hidden in massive data, including all human RNAseq data from SRA database. We then generated quantitative patterns from this network to uncover distinctive mechanisms of ulncRNA activation, regulation, and function.
2. Materials and methods
2.1. RNAseq data resource and download
We downloaded all human RNAseq data from Sequence Read Archive (SRA) as we previously described [19]. Briefly, this study searched Homo sapiens and RNA_seq from the SRA database and got a total of 265,361 SRA sample IDs containing various experimental conditions such as tissues and cell lines (Table_S1). All detailed info is available on our project website [20]. LncRNA endogenous in this data set should be endogenous in the human genome. The SRR number (SRR#) for each sample extracted from these IDs was used to prefetch its sra format files via sratoolkit.2.8 [21]. The sra file was converted to fastq file via fastq-dump 2.8 from the same package of sratoolkit.
2.2. Alignment
The fastq files were aligned to GRCh38.p10.v27 by using STAR-2.5 [22] with the following settings: runThreadN 30 --genomeDir GRCh38.p10.v27 --outSAMtype BAM Unsorted SortedByCoordinate --outFilterMultimapNmax 20 --outFilterType BySJout --chimSegmentMin 20 --alignSJoverhangMin 8 --alignSJDBoverhangMin 1 --quantMode TranscriptomeSAM GeneCounts --outFilterIntronMotifs RemoveNoncanonical --twopassMode Basic.
All 63,925 unique genes annotated by GRCh38.p10.v27 were used to count gene read depth by STAR as running above. The aligned BAM files were used to count read depth for ulncRNAs, in which unannotated regions (≥10 bp distance from annotated regions) were split into 300 bp fragments as putative pre-ulncRNAs to count reads (see main text and Fig. 1A). This 300 bp was used as the basic unit because lncRNA length is normally defined as longer than 200 bp [6].
Fig. 1.
Functionally endogenous ulncRNAs identified in the human genome. A, A workflow of ulncRNA regulatory network identification. Green, orange and blue bars represent three genetic fragments, and 1, 2, 3 and 4 denote four 300 bp spit fragment labels. B, a sample network of an ulncRNA (ID: chr3.36642256.36642555). ulncRNA ID was named as chromosome plus coordinate. Node label color denotes the gene category, red:protein, green:ulncRNA, purple:annotated noncoding RNAs. Edge color represents significance (p-value), from low (yellow) to dark blue (high, low p-value) inferred by FINET. Edge thickness denotes confidence measured by frequency score in our FINET software, thicker, more confident. C, Overlap of lncRNAs from FANTOM and total 16,594 unique ulncRNAs identified by this present study via using FINET software. D, size distribution of total 16,594 ulncRNAs. E, Distribution of minimum distance from ulncRNAs to proteins. F, ulncRNA density along chromosomes measured by total length of RNAs/chromosome length (Fig. S2 for detail). G, an example profiling of ulncRNA, chr19:58381218.58384517. Its log2 expression level in TPM was shown in brown, and the profiling of two typical histone markers was plotted, H3K4me1(ENCODE ID: ENCFF730CTY.bigWig) and H3K4me3 (ENCODE ID: ENCFF881MFX.bigWig). Annotated genes ZNF837, RIPS5, MIR4754 were marked at the bottom. H, gene category proportion of top 1000 centrality in ulncRNA network. P_ denotes processed. For example, p_pseudogene as processed_pseudogene. I, top 10 highest connected nodes in ulncRNA network. Frequency count represents interactions (degree). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Read counting was performed by htseq-count in HTSeq 0.12.3 [23] with no strand-specific.
2.3. Sample filtering
We focused on high-quality samples with the whole transcriptome, and automatically filtered out any abnormal samples. We first filtered out any abnormal samples from downloaded and aligned steps, such as unauthorized, unpublic, undownloadable, unaligned to the whole transcriptome, and uncountable for the whole whole transcriptome. The last two represented non-whole transcriptomes. These filtered steps generated 65,314 samples from 265,361.
Many genes have a zero read count in RNAseq. We further filtered out samples with zero counts in most annotated genes and filtered out samples containing >50,000 genes with zero count as done in our paralleled study [19]. This 50,000 zero count cut-off was based on zero distribution in control RNAseq of The Cancer Genome Atlas (TCGA) data [24], which was a large set (>11000 samples) of high quality data with whole transcriptome generated by unified standard protocol [19]. Finally we got 26,896 high quality samples for the rest of the analysis (Table_S2).
2.4. Calculating TPM and filtering genes
To ensure gene expression comparable for each sample, we normalized RNAseq data by calculating transcripts per million (TPM) for each sample as following:
TPM = ratio / sum(ratio) * 1,000,000
ratio = read counts /gene lengths
The gene length of annotated genes was defined by GENCODE project [7], and the ulncRNA length was the range of a ulncRNA coordinate.
After TPM calculating, we performed gene filtering and counted zero TPM for each annotated gene throughout all samples. Genes with zero counts across all samples were removed and only genes with nonzero counts >3 were kept for downstream analysis. Final 58,871 annotated genes were kept in 26,896 SRA samples.
Normally, ulncRNAs express low and carry more zero counts. Thus, we filtered ulncRNAs with zero TPM >5000 based on zero distribution of ulncRNAs and finally got 116,678 pre_ulncRNAs to build a regulatory network as described below.
2.5. Construction of the lncRNA endogenous regulatory network
The filtered data for both annotated genes and pre_ulncRNAs constructed a TPM matrix, in which rows and columns were respectively corresponding to samples and genes. This TPM matrix was used to build a regulatory network by using our software dubbed FINET [18]. FINET treats each gene as a target (Y) in the matrix containing 58,871 annotated genes and 116,678 pre_ulncRNAs and searches its regulators from the rest of genes (X) to build a direct network via elastic-net. This elastic-net-based search could contain 90% of false positive interactions [18]. To reduce this type of error, FINET introduces stability-selection [25] and randomly splits the samples into m sub-groups (m = 8 in this study, 3362 samples in each subgroup) and then searches target-regulator interactions from each sub-group. If an interaction consistently occurs in m sub-groups, the type I error is very low [18], [25] and this error dramatically reduces when m value becomes large in complex biological data [18]. This stability-selection repeats n times (n = 50 in this study). A frequency score (frequency in m*n trials) was calculated for each target-regulator interaction. A perfect frequency score (frequency score = 1) represents that an interaction always occurs (100%) in m*n random trials. This study used frequency score 0.95 (p = 0.95 as shown in a running command line below) as cutoff to filter out interactions. A large number of regulatory interactions with low frequency scores were normally specific for certain conditions like a given experimental cell type and these interactions were filtered out at this step. The leftover interactions occurred >380 times out of 400 trials and were treated as endogenous target-regulator interactions independent of conditions. These endogenous interactions constructed a lncRNA endogenous regulatory network in the human genome.
We run FINET as: julia finet.jl -c 120 -k 5 -n 50 -m 8 -a 0.5 -p 0.95 -i TPM_matrix -o mynetwork
The output network was deposited in our server [20].
We concatenate conjunctions of pre-ulncRNAs if they are close neighbors with distance of 1 bp (e.g. concatenate 0–299 to 300–599 to 0–599).
2.6. Network centrality
Network centrality was calculated by using NetworkX2.5 implemented in python3.8 [26]. To avoid biases, we calculated two types of centrality, degree and eigenvector. Genes with degree and eigenvector centrality were ranked on the basis of ranking score as approached for network node ranking [27]. The final ranking was generated on the basis of the sum of two ranking scores as practiced in gene ranking in a regulatory network [27]. The highest ranked nodes were treated as network hubs.
The degree (frequency count) of a network node was counted by its total interactions.
2.7. Module identification and category
To understand ulncRNA functions, we searched ulncRNA target modules in which ulncRNAs as regulators and protein coding genes as targets. We filtered the entire network with ulncRNAs as regulators and protein coding genes as targets and got a sub-network of ulncRNA targeting protein coding genes. The modules of this sub-network were searched by spectral partitioning algorithm via using MODULAR Alfa 0.21 under Linux terminal [28], [29]. MODULAR was designed to facilitate module identifications from nature networks through maximization of the degree of modularity. Implemented by C language, MODULAR computes fast and autonomously when detecting modules. We run MODULAR by inputting a unipartite network in UCINET edgelist format and then optimizing by spectral partitioning.
2.8. ulncRNA transcription initiation
Human transcription marker binding data were downloaded in both bed and bam files from ENCODE [2]. All files aligned to GRCh38 were selected or converted to GRCh38 by LiftOver [30]. A total of 780 peak bed samples containing top 9 abundant measurements of transcription initiation were downloaded, including 104, 102, 51, 182, 14, 110, 111, and 95 of Chip-seq peak bed files respectively for H3K4me1, H3K27ac, H3K9ac, H3K4me3, POLR2A, H3K36me3, H3K27me3, H3K9me3, and 11 ATAC_seq bed files (Table_S3).
We examined the transcription marker binding abundance in the ulncRNA putative promoter region, which was defined as 5000 bp upstream from ulncRNA transcription start site (TSS) that is the first base pair of either 5′ or 3′ in the genome coordinate of a ulncRNA. We counted the binding peaks of either 5′ or 3′ of a lncRNA but only count one peak even if more binding peaks might present at either end (5′ or 3′) for a given marker and a given sample.
In addition, a total of 1918 and 2694 bam files respectively for H3K4m1 and H3K4me3 were also downloaded to compute the promoter matrix (Table_S3).
Transcription marker measurements varied with tissues and cell lines in the ENCODE project. For unbiased results, we did not filter out any tissues and cell types and included all measurements conducted by ENCODE.
2.9. Statistics
All statistics analysis and presentation were performed by R 3.6 libraries including ggplot2, ggVennDiagram and GenomicRanges. All labeled p-values were derived from t test unless specifically noted in this study. Network was visualized by cytoscape 3.7 [31].
3. Results
3.1. Endogenous ulncRNA regulatory network
The data from our previous study revealed that only 22% of lncRNAs annotated by the GENCODE project are functionally endogenous (Fig. S1) [19], indicating that most annotated lncRNAs are tissue-specific. The number of endogenous ulncRNAs in dark regions of the human genome remains unknown.
To identify endogenous ulncRNAs, we used TPM as expression value (materials and methods) and developed an algorithm strategy to systematically capture all functional ulncRNAs endogenous across all human tissues and conditions (Fig. 1A). This strategy includes the following 4 key steps. (1) split unannotated dark regions (≥10 bp distance from annotated regions) into 300 bp RNA fragments as preliminary ulncRNAs (Fig. 1A, materials and methods). (2) identify interactions of ulncRNAs endogenous in human genome from massive data (all RNAseq data deposited in SRA) by using our FINET software that infers endogenous regulatory interactions from massive data with high accuracy [18] via integrating algorithms with stability-selection, elastic-net, and parameter optimization (materials and methods), and simultaneously remove nonfunctional ulncRNAs with no regulatory interactions (e.g. removing the blue color ulncRNA in Fig. 1A). (3) concatenate conjunctions (e.g. concatenate ulncRNA1 and ulncRNA2 into ulncRNA12 in Fig. 1A). (4) assemble all interactions into a ulncRNA regulatory network, in which an individual ulncRNA possesses at least one functionally regulatory interaction. This strategy generated an endogenous ulncRNA regulatory network, which includes a final set of 16,594 active unique ulncRNAs with 62,586 edges and 29,794 nodes (Fig. 1A–B). This whole network was deposited and searchable from the project website [20].
Among 16,594 ulncRNAs, only 4.5% (758/16594) overlapped with lncRNAs identified by FANTOM project (Fig. 1C), which used experiments to identify lncRNAs. Biological experiments are usually performed by using specific conditions like specific cell types and results derived from these experiments could be biased to experimental conditions. In contrast, our results come from big data from various conditions and results are unbiased. This 4.5% overlap indicated that most of lncRNAs identified by both the FANTOM and GENCODE projects are cell type specific.
Overall, we revealed a novel ulncRNA regulatory network endogenous across human genomes and conditions deposited in the SRA database, with no cell type specificity.
3.2. Key characteristics of endogenous ulncRNAs
To understand the primary characteristics of these 16,594 ulncRNAs, we examined the distribution of their length, closest distance to protein-coding sequences, chromosome distribution, and key hubs in the network. Most of ulncRNAs (65%) were <600 bp length (43% of 600 bp + 22% 300 bp) (Fig. 1D), and long ulncRNAs (>1500 bp) occupied 12%. Surprisingly, most of these ulncRNAs distribute far away from protein-coding regions, with >62% located >5 kb from protein-coding regions (Fig. 1E). We also calculated the ulncRNA density across chromosome (total ulncRNAs length/chromosome length) and found that chr19 possessed the most density of ulncRNAs, with >1 bp ulncRNAs in every 100 bp DNA length (Fig. 1F, Fig. S2). An example in chr19 (id:chr19.58381218.58384517) was shown in Fig. 1G.
To understand the crucial hubs in the ulncRNA network, we examined both the centrality of the entire network and the highest connected nodes. ulncRNAs worked as the key hubs in this network and they occupied >50% of top 1000 centrality and 90% of top 50 as ulncRNA centrality (Fig. 1H, materials and methods). In addition, 8 out of top 10 highest connected nodes were ulncRNAs (Fig. 1I). The top 1 of these ulncRNAs, chr13.109423936.109424535, connected with total 5511 components in the entire network. This indicated that ulncRNAs, rather than protein-coding genes, predominate the network hubs and degrees, suggesting ulncRNAs govern the entire ulncRNA network and ulncRNA regime, instead of protein coding genes.
Together, these above results suggested that the ulncRNA regime is overall distinctive from that of protein-coding genes, in which ulncRNAs are short, far away from coding regions, varied in chromosome distribution, and controlled by ulncRNA themselves.
3.3. Systematic mechanisms of ulncRNA transcription initiation
The mechanisms of lncRNA initiation have been intensely debated [6]. Protein-based mechanisms such as enhancers and RNA polymerase II have been thought as the primary factors for lncRNA activation [6]. To understand whether the protein-based mechanisms can be applied to ulncRNA initiation, we systematically compared the binding distributions of transcription markers of ulncRNAs and protein-coding genes (materials and methods). To make binding profiles comparable between protein-coding genes and ulncRNAs, we used the same number of ulncRNAs and protein-coding genes. From our previous study, we learned that 14,122 protein coding genes were active in normal conditions [19], thus we randomly selected 14,122 ulncRNAs out of 16,594 to match the protein number. Total 9 factors that are abundantly measured by ENCODE were examined, including ATAC_seq [32], [33], 3 markers for enhancer (H3K4me1, H3K27ac, H3K9ac) [34], [35], [36], 3 for promoter (H3K4me3, POLR2A, H3K36me3) [37], [38], and 2 for silence and tissue specificity (H3K27me3 and H3K9me3) [6], [39].
We counted the binding peak frequency of each factor across promoter regions of these 14,122 ulncRNAs (materials and methods). Surprisingly, POLR2A barely bound to lncRNA promoter regions and it bound only 1668 (median) out of 14,122 ulncRNAs (Fig. 2A), indicating that >88% of ulncRNAs were not associated with POLR2A during their initiations. This suggested that polymerase II plays much less role than previously thought in activating ulncRNA transcription.
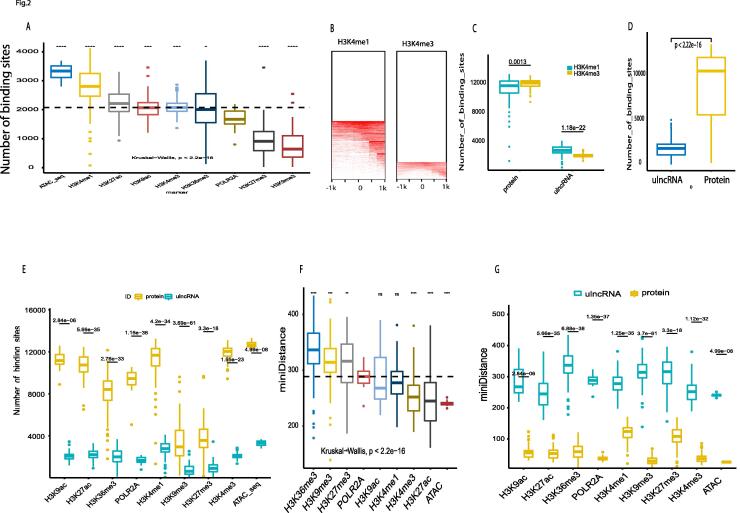
Fig. 2.
Contributions of histone and transcription marker to the ulncRNA activation. A, the frequency (total number of binding sites) of 9 measurements (8 markers and ATAC_seq) that bind to ulncRNA promoter regions (within 1000 bp from TSS, transcription start site). The black line represents the median of POLR2A binding sites (1668). These 9 measurement data were extracted from ENCODE database (https://combai.org/static/ids/ulncRNA_encodesamples.zip). Significance level, **,**** denotes p value <0.0034 and 3.4e-06 respectively. B, binding heatmap of H3K4me1 and H3K4me3. The heatmap was plotted by using the median of 1918 and 2694 samples respectively for H3K4me1 and H3K4me3 measured by the ENCODE project (H3K4me1bam and H3K4me3bam). C, Comparison H3K4me1 and H3K4me3 binding sites of ulncRNAs and protein coding genes. Labeled number represents p value derived from t test in this study. D, Comparison of total binding sites of 9 measurements between ulncRNAs and proteins. E, Binding comparison of each measurement between ulncRNAs and proteins. F, minimum distance (bp) from factor binding to TSS. **,***,**** denote p < 0.0055, 0.00053, and 5.7e-08 respectively. G, minimum distance (bp) comparison of each factor between ulncRNAs and proteins.
In contrast, all three enhancer biomarkers, H3K4me1, H3K27ac, and H3K9ac, exhibited significantly higher binding sites than POLR2A (Kruskal–Wallis, p < 2.2e − 16, Fig. 2A). Furthermore, H3K4m1 bindings were significantly higher than H3K4me3, a marker for active promoters near TSS, (Fig. 2B-C, Fig. 1G), while H3K4me3 bindings were higher in protein coding genes (Fig. 2C). This indicates that enhancers play a much greater role than polymerase II in activation of ulncRNAs.
To better understand the whole picture of these factor bindings, we box-plotted all the binding sites for ulncRNAs and protein coding genes. ulncRNAs contained significantly lower bindings than protein coding genes (Fig. 2D–E), at median of 1990 and 10,321 for ulncRNAs and protein coding genes respectively (Fig. 2D), indicating that only 14% (1990/14122) of ulncRNAs possessed one biomarker to bind while 73% (10321/14122) of proteins have at least one biomarker to bind. Actually, all these histone marker bindings to ulncRNAs were significantly lower than to protein coding genes (Fig. 2E, Fig. S3). For example, H3K4me3, H3K4me1 and POLR2A densely bound to protein coding genes with 85% (12035/14122), 83% (11666/14122) and 67% (9446/14122) respectively (Fig. S3). Moreover, protein coding genes simultaneously possessed multiple factors for enhancers (e.g. ACTA and H3K4me1) and promoters (H3K4me3) to densely bind (Fig. 2E), but ulncRNAs possessed much fewer factors to bind. This might partially interpret the low expression level of ulncRNAs. Moreover, the overall low bindings and the low H3K4m1 bindings were obviously not sufficient to activate the widespread ulncRNAs, suggesting that the key mechanism accounting for the majority of ulncRNAs activation remains to be investigated.
To appreciate the binding distance to ulncRNA TSS, we calculated the minimum distance from factor bindings to TSS. The minimum distance median ranged from 240pb (ATAC) to 336 bp (H3K36me3) (Fig. 2F). Factors with most binding sites (Fig. 2A), including ATAC, H3K4me1, H3K27ac, H3K9ac, and H3K4me3, were bound to ulncRNAs with short distance to TSS (Fig. 2F). However, these short distances for ulncRNAs were significantly longer than protein coding genes, in which factors bound much closer to protein coding gene TSS (Fig. 2G, Fig. S4, Fig. S5). Furthermore, four markers (ATAC, H3K9me3, H3K4me3, POLR2A) were bound to protein coding gene TSS within 50 bp (median) while the rest within 120 bp (median) (Fig. S4). In contrast, the median for all ulncRNAs was close to 280 bp (Fig. S5). This was another line of evidence for ulncRNA initiation regions distinctive from protein coding genes. Altogether, these above suggested that ulncRNA activation mechanism is different from protein coding genes and that the alternative mechanism for ulncRNA activation remains dark.
3.4. ulncRNA regulators
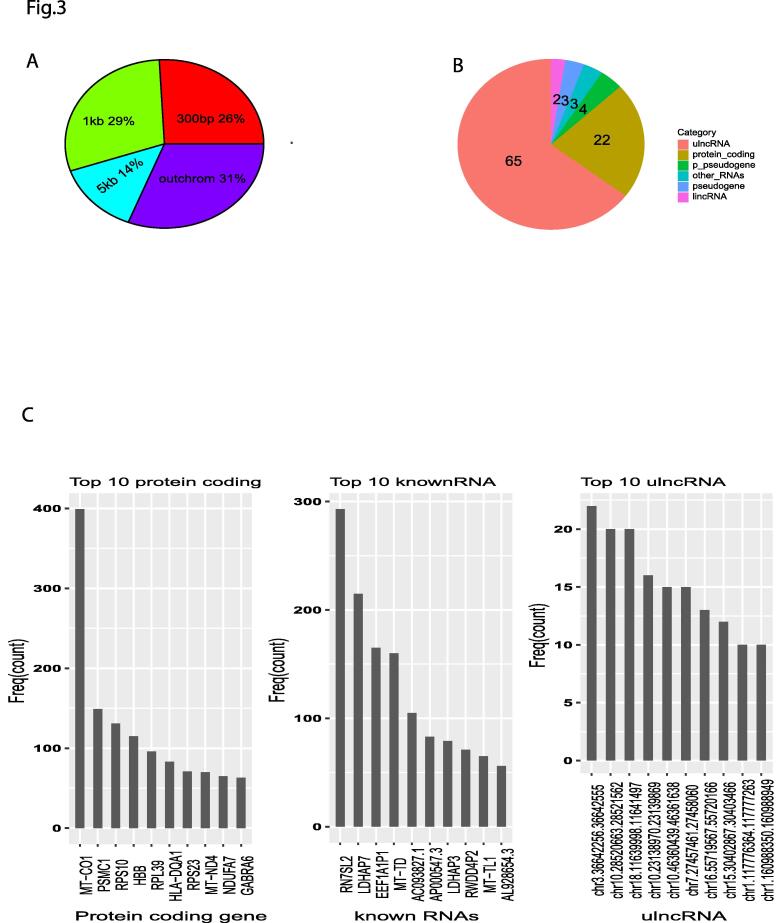
Our network was a direct regulatory network (materials and methods), in which a regulator was defined as a node that directly points to a targeting node (gene). To find regulators for a ulncRNA, we treated this ulncRNA as a target and found all regulators pointed to this ulncRNA. We searched all regulators of 16,594 lncRNAs in the entire ulncRNA regulatory network and found a total of 31,051 genes regulating ulncRNAs. We examined the genome coordinates of all these regulators and found that the most abundant regulators (>31%) were located outside their own chromosomes (Fig. 3A), suggesting that almost a third of ulncRNAs are trans-regulated. In addition, 65% of these 31,051 regulators were ulncRNAs (Fig. 3B), suggesting that ulncRNAs primarily regulate themselves, consistent with our previous studies showing that noncoding genes tend to trans-regulate themselves in the same category [19].
Fig. 3.
ulncRNA Regulators. A, distribution of distance from ulncRNA regulators to ulncRNAs (percentage of total 31,051 ulncRNA regulators). B, gene categories of ulncRNA regulators (% of total 31,051 ulncRNA regulators). C, top 10 highest connected regulators of protein coding genes, annotated RNAs, and ulncRNAs.
In contrast to the conventional notion that proteins serve as primary regulators for lncRNAs, proteins actually work as secondary regulators (22%) for ulncRNAs (Fig. 3B). Among protein regulators, a mitochondrial protein MT-CO1 connected to most ulncRNAs, with 400 interactions (Fig. 3C left panel). Another mitochondrial protein MT-ND4 was also ranked as top 8 highest connected regulators (Fig. 3C left panel). Moreover, two annotated noncoding RNAs, MT-TD and MT-TL1, were also in top 10 noncoding regulators for ulncRNAs (Fig. 3C right). This suggested that mitochondrial components play a critical role in regulating ulncRNAs.
3.5. ulncRNA targets
Whether lncRNAs target their neighbor genes is debated [6], [12]. We plotted gene expression regression of ulncRNAs and their closest proteins within distance of 300 bp and 1000 bp respectively, and found that ulncRNAs did not regulate their neighbor protein coding genes (Fig. 4A, Fig. S6). Instead ulncRNAs regulate their targets in a trans-regulatory manner, with the majority of ulncRNAs (57%) across chromosomes (Fig. 4B). This parallels with a recent observation showing that the majority of lncRNAs are located in cytoplasm as trans-regulators [40]. This is also consistent with our study on annotated lncRNA trans-regulation mechanisms [19].
Fig. 4.
ulncRNA targets. A, Gene expression correlation between ulncRNAs and their closest proteins (within 300 bp). B, Distribution of distances from ulncRNAs to their targets (percentage of total 51,633 ulncRNA targets). C, gene categories of ulncRNA targets (% of total 51,633 ulncRNA targets). D, top 10 highest connected ulncRNA targets of protein coding genes, annotated RNAs, and ulncRNAs. UlncRNA targets were ranked by their degree (freq count) from the entire network. E, top 4 functional modules in ulncRNA network. The size of the module represents its member abundance. F, Functionally scheme of ulncRNA. The arrow size and line thickness represent the quantitative weight of importance. For ulncRNA activation, the factor importance ranking is as follows: unknown factor > ulncRNA > histone > POL II.
The majority of ulncRNAs target protein coding genes (55%, Fig. 4C), indicating their broad regulatory role. However, their targets were thinly scattered. The top 1 protein coding gene target (ARF6) ranked by network degree only carries 12 interactions and the top noncoding RNAs and ulncRNAs have <10 interactions (Fig. 4D). Comparing hundreds of protein coding gene targets [19], ulncRNAs regulate their targets in a fine way. Together, these above suggested that ulncRNAs primarily perform broad- but fine-regulation toward their targets.
3.6. ulncRNA primary functions
To understand the primary functions of ulncRNAs, we investigated the key biological functions of ulncRNA targets. Among ulncRNA targets, protein coding genes dominated the whole profiling (>55%, Fig. 4C) and their functions should represent the primary functions of ulncRNA targets. We searched ulncRNA target modules by spectral partitioning algorithm [28], [29]. Total 154 modules were generated, but only 4 of them were functionally enriched. The rest were of small member size (member number < 5) (Fig. 4E, Figs. S7–S10). Their functions were primarily relevant to RNA processes but included 7 key categories,1) transcription and RNA processing (RNA splicing, ncRNA metabolic and processing); 2) mitotic cell cycle and DNA replication; 3) cellular respiration; 4) cellular response to stress (DNA repair); 5) chromatin organization; 6) translation and post- translational protein modification, proteasomal protein catabolic process, and protein localization; 7) nervous system development. These ulncRNA target functions suggested that biological roles of ulncRNAs are broad.
To summarize, histone modifications on enhancers play a more important role in activating ulncRNAs than polymerase II in promoter regions, but these histone modifications only account for a small proportion (<20%) of ulncRNA originations. The ulncRNA self-regulation and unknown mechanism activate the large proportion of ulncRNAs, resulting in an array of bioprocess activation (Fig. 4F).
4. Discussion
This present study systematically decoded functionally endogenous lncRNAs from unannotated regions of the human genome and revealed a functionally distinctive regime for lncRNAs. LncRNAs are widely expressed throughout the human genome, but only a small proportion of lncRNAs have been identified and these annotated lncRNAs are mostly tissue specific [8], [9]. Little has been known about lncRNA endogeneity in the human genome. This present study overcame the limitations of tissues and conditions by using all RNAseq data from SRA and revealed 16,594 endogenous lncRNAs in the human genome. These lncRNAs mostly self-regulate independently of protein coding genes and establish their own functional regime distinctively from proteins in terms of distribution, activation, regulation and function.
The mRNA initiation concept has been widely applied to lncRNA study [6]. Several mechanisms have been proposed for lncRNA initiation, such as promoter and POL II and protein-based histone modifications on enhancers [6], [8]. However, our data revealed that these conventional mRNA-based mechanisms only account for around 20% of lncRNA activation. lncRNA self-regulation generally contributes to their activation because individual lncRNA expressions are heavily regulated by other individual ulncRNAs and lncRNA expression levels primarily result from lncRNA-self regulation. In normal conditions, these regulations stay weak to save energy, but under stimulation a certain group of lncRNAs would be highly activated by an array of lncRNA individuals and perform their biology functions [3], [19]. The complete big picture of lncRNA activation independent on protein-based polymerase remains to be further investigated.
lncRNAs are expressed at much lower levels than mRNAs. The mechanism for that remains debated [6]. The short life-span and the low promoter transcription efficiency have been thought as the mechanism of low lncRNA expression, but recent studies have demonstrated that lncRNAs have a similar half-life as normal mRNA and the bi-directional promoter working for protein coding genes perform similarly for lncRNAs [6]. These two mechanisms hardly interpret the low lncRNA expression. The critical mistakes in these two mechanisms resulted from an assumption that lncRNAs employ the same mechanisms of protein-based promoters. Our data showed that ulncRNAs rarely use protein-based promoter mechanisms but they partially employ enhancers far away from normal protein coding gene promoters. This parallels recent observations showing lncRNA initiations from enhancers [9]. However, this enhancer initiation for lncRNAs is distinctive from protein coding genes. In protein coding gene regime, histone modifications like H3K4me2 are sufficient for transcription initiation, but all these protein-based factor bindings to lncRNAs are too low to initiate widespread lncRNAs regardless of enhancers and promoters. These low bindings of all transcription markers at least interpret the partial mechanism of low lncRNA expression.
lncRNAs were once thought of as junk with no functions, but recently their functions have been recognized as regulators in several important processes such as growth and metabolism [4], [41], [42], [43]. Our recent study also unearthed noncoding RNAs as the universal deadliest regulators for all cancers [3]. However, the primary functions of the vast majority of human genome occupied by lncRNAs still remain unknown. Here, we systematically reveal they target protein coding genes functioning in an array of bioprocesses, such as transcription and RNA processing, mitotic cell cycle and DNA replication. These help us to understand the basic mechanism of lncRNA biological functions.
LncRNAs pre-dominate most of the human genome and have their own regime distinct from proteins. Applying protein coding gene strategy and concept to understand lncRNAs may be misleading. We need to create a novel concept system to understand lncRNAs and the human functional genome.
It is challenging for both biologists and computational scientists to dig out the big picture from massive biological data due to strong background noise and mixed labels in the database. This present study employed FINET to systematically remove noise and condition effects and revealed the endogenous human ulncRNAs from huge data. This established a new avenue to unearth biological meaningful patterns from increasing massive data. We recently have applied the similar algorithm and deep learning neural networks to identify universal biomarkers for detecting cancers [44]. Combining FINET and artificial intelligence algorithms helps to accelerate novel discoveries in this big data era.
Acknowledgments
Acknowledgements
The data were downloaded from Sequence Read Archive (SRA) and The Cancer Genome Atlas (TCGA).
Funding source
No funding associated with the project.
Declaration section
Data and material availability.
Data deposited in the project website [20]
Competing interests
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.05.020.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447:799–816. [DOI] [PMC free article] [PubMed]
- 2.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang A., Hai R. Noncoding RNAs Serve as the Deadliest Universal Regulators of all Cancers. Cancer Genomics Proteomics. 2021;18:43–52. doi: 10.21873/cgp.20240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wei L.-H., Guo J.U. Coding functions of “noncoding” RNAs. Science. 2020;367:1074–1075. doi: 10.1126/science.aba6117. [DOI] [PubMed] [Google Scholar]
- 5.Ramilowski J.A., Yip C.W., Agrawal S., Chang J.-C., Ciani Y., Kulakovskiy I.V., et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 2020;30:1060–1072. doi: 10.1101/gr.254219.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ransohoff J.D., Wei Y., Khavari P.A. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19:143–157. doi: 10.1038/nrm.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.GENCODE - Human Release 35, https://www.gencodegenes.org/human/release_35.html n.d.
- 8.Schlackow M., Nojima T., Gomes T., Dhir A., Carmo-Fonseca M., Proudfoot N.J. Distinctive Patterns of transcription and RNA processing for human lincRNAs. Mol Cell. 2017;65:25–38. doi: 10.1016/j.molcel.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hon C.-C., Ramilowski J.A., Harshbarger J., Bertin N., Rackham O.J.L., Gough J., et al. An atlas of human long non-coding RNAs with accurate 5′ ends. Nature. 2017;543:199–204. doi: 10.1038/nature21374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Musacchia F., Basu S., Petrosino G., Salvemini M., Sanges R. Annocript: a flexible pipeline for the annotation of transcriptomes able to identify putative long noncoding RNAs. Bioinformatics. 2015;31:2199–2201. doi: 10.1093/bioinformatics/btv106. [DOI] [PubMed] [Google Scholar]
- 11.Lin M.F., Jungreis I., Kellis M. PhyloCSF: a comparative genomics method to distinguish protein coding and non-coding regions. Bioinformatics. 2011;27:i275–i282. doi: 10.1093/bioinformatics/btr209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cabili M.N., Trapnell C., Goff L., Koziol M., Tazon-Vega B., Regev A., et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25:1915–1927. doi: 10.1101/gad.17446611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guttman M., Amit I., Garber M., French C., Lin M.F., Feldser D., et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chinwalla A.T., Cook L.L., Delehaunty K.D., Fewell G.A., Fulton L.A., Fulton R.S., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 15.Howe K., Clark M.D., Torroja C.F., Torrance J., Berthelot C., Muffato M., et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 2013;496:498–503. doi: 10.1038/nature12111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin J.T., Priest J.R., Ovcharenko I., Ronco A., Moore R.K., Burns C.G., et al. Human-zebrafish non-coding conserved elements act in vivo to regulate transcription. Nucleic Acids Res. 2005;33:5437–5445. doi: 10.1093/nar/gki853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.NONCODE n.d. http://www.noncode.org/ (accessed May 5, 2022).
- 18.Wang A., Hai R. FINET: Fast Inferring NETwork. BMC Res Notes. 2020;13:521. doi: 10.1186/s13104-020-05371-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang A. Noncoding RNAs endogenously rule the cancerous regulatory realm while proteins govern the normal. Comput Struct Biotechnol J. 2022;20:1935–1945. doi: 10.1016/j.csbj.2022.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang A. ulncRNA network, https://combai.org/network/lncRNA/. n.d.
- 21.SRA Toolkit – SBGrid Consortium – Supported Software n.d. https://sbgrid.org/software/titles/sra-toolkit (accessed January 25, 2022).
- 22.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., et al. STAR: ultrafast universal RNA-seq aligner. Bioinforma Oxf Engl. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anders S., Pyl P.T., Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomczak K., Czerwińska P., Wiznerowicz M. The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol Poznan Pol. 2015;19:A68–A77. doi: 10.5114/wo.2014.47136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinshausen N., Bühlmann P. Stability selection. J R Stat Soc Ser B Stat Methodol. 2010;72:417–473. doi: 10.1111/j.1467-9868.2010.00740.x. [DOI] [Google Scholar]
- 26.Proceedings of the Python in Science Conference (SciPy): Exploring Network Structure, Dynamics, and Function using NetworkX n.d. http://conference.scipy.org/proceedings/SciPy2008/paper_2/ (accessed December 31, 2021).
- 27.Marbach D., Costello J.C., Küffner R., Vega N.M., Prill R.J., Camacho D.M., et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newman M.E.J. Finding community structure in networks using the eigenvectors of matrices. Phys Rev E. 2006;74 doi: 10.1103/PhysRevE.74.036104. [DOI] [PubMed] [Google Scholar]
- 29.Marquitti F.M.D., Guimarães P.R., Pires M.M., Bittencourt L.F. MODULAR: software for the autonomous computation of modularity in large network sets. Ecography. 2014;37:221–224. doi: 10.1111/j.1600-0587.2013.00506.x. [DOI] [Google Scholar]
- 30.Lift Genome Annotations n.d. http://genome.ucsc.edu/cgi-bin/hgLiftOver (accessed March 16, 2022).
- 31.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Landt S.G., Marinov G.K., Kundaje A., Kheradpour P., Pauli F., Batzoglou S., et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Domcke S., Hill A.J., Daza R.M., Cao J., O’Day D.R., Pliner H.A., et al. A human cell atlas of fetal chromatin accessibility. Science. 2020;370 doi: 10.1126/science.aba7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rada-Iglesias A. Is H3K4me1 at enhancers correlative or causative? Nat Genet. 2018;50:4–5. doi: 10.1038/s41588-017-0018-3. [DOI] [PubMed] [Google Scholar]
- 35.Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karmodiya K., Krebs A.R., Oulad-Abdelghani M., Kimura H., Tora L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genomics. 2012;13:424. doi: 10.1186/1471-2164-13-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang G., Lin J.C.Y., Wei V., Yoo C., Cheng J.C., Nguyen C.T., et al. Distinct localization of histone H3 acetylation and H3–K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolasinska-Zwierz P., Down T., Latorre I., Liu T., Liu X.S., Ahringer J. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet. 2009;41:376–381. doi: 10.1038/ng.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 40.Carlevaro-Fita J., Rahim A., Guigó R., Vardy L.A., Johnson R. Cytoplasmic long noncoding RNAs are frequently bound to and degraded at ribosomes in human cells. RNA. 2016;22:867–882. doi: 10.1261/rna.053561.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ramilowski J., Yip C.W., Agrawal S., Chang J.-C., Ciani Y., Kulakovskiy I.V., et al. Functional annotation of human long non-coding RNAs via molecular phenotyping. BioRxiv. 2019;700864 doi: 10.1101/700864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parenteau J., Maignon L., Berthoumieux M., Catala M., Gagnon V., Abou E.S. Introns are mediators of cell response to starvation. Nature. 2019;565:612–617. doi: 10.1038/s41586-018-0859-7. [DOI] [PubMed] [Google Scholar]
- 43.Morgan J.T., Fink G.R., Bartel D.P. Excised linear introns regulate growth in yeast. Nature. 2019;565:606–611. doi: 10.1038/s41586-018-0828-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang A., Hai R., Rider P.J., He Q. Noncoding RNAs and deep learning neural network discriminate multi-cancer types. Cancers. 2022;14:352. doi: 10.3390/cancers14020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.