Abstract
This study is the largest analysis of DNA viruses in solid tumors with associated genomics. To achieve this, a novel method for discovery of DNA viruses from matched tumor/normal next-generation sequencing samples was developed and validated. This method performed comparably to reference methods for the detection of high-risk (HR) human papilloma virus (HPV) (area under the receiver operating characteristic curve = 0.953). After virus identification in 48,148 consecutives samples from 42,846 unique patients, novel virus tumor associations were established by segregating tumor types to determine whether each DNA virus was enriched in each of the tumor types compared with the remaining cohort. All firmly established solid tumor-virus associations (eg, HR HPV in cervical cancer) were confirmed, and the novel associations discovered included: human herpes virus 6 in neuroblastoma, human herpes virus 7 in esophagogastric cancer, and HPV42 in digital papillary adenocarcinoma. These associations were confirmed in an independent validation cohort. HR HPV– and Epstein-Barr virus–associated tumors showed newly discovered genomic associations, including a lower tumor mutation burden. The study demonstrated the ability to study the role of DNA viruses in human cancer from clinical genomics data and established the largest cohort that can be utilized as a validation set for future discovery efforts.
Viruses are among the most well-established causal agents for cellular transformation to malignancy in solid tumors. The potential for viruses to transform healthy cells to malignant solid tumors was first described in 1911 by Rous1 in an animal model. The ability of viruses to induce human tumors was established by investigators in a variety of solid tumors later in the century.2, 3, 4, 5, 6, 7, 8, 9 The most recent description of a causal association between a virus and a solid tumor was the establishment of Merkel cell polyoma virus as the causal agent in Merkel cell carcinoma. Merkel cell polyoma virus was discovered in 2008 with next-generation sequencing (NGS) technology and a digital subtraction method to remove human genome reads from the analysis of RNA sequencing reads to detect viral RNA.10
With the advent of large-scale genome sequencing projects, the opportunity to investigate virus-tumor associations has emerged by examining various data sources: RNA sequencing, whole genome sequencing, and whole exome sequencing; sources available through The Cancer Genome Atlas Research Network.11, 12, 13, 14 The ability to apply a similar digital subtraction technique, whereby sequencing reads that do not align to the human genome are used to identify presence of viral DNA, has not previously been investigated in the context of clinical genomics. This presents the opportunity for analysis of the viral status for large cohorts of tumors. To date, investigators have yet to demonstrate the feasibility and validate the performance of such an analysis in clinical sequencing data.
The current study introduces a technique for virus detection and discovery in clinical, hybrid capture-based, large-panel NGS data and perform a proof-of-principle analysis in a clinical sequencing cohort of 48,148 solid tumor samples. It shows that without performing additional sequencing, virus-tumor type associations can be interrogated with high accuracy and fidelity. It explores the genomic correlates of virus-associated tumors and that this viral data can immediately inform diagnosis and patient management.
Materials and Methods
Data Set
This study was completed utilizing data generated for the Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) clinical assay, a US Food and Drug Administration–cleared tumor profiling assay for patients with advanced solid cancers. MSK-IMPACT Solid is optimized for the detection of clinically relevant somatic mutations in patients with solid tumors.15,16 The study was approved by the institutional review board (Memorial Sloan Kettering Cancer Center; 12-245 and 18-128). The data used for this study are based on the first 48,148 solid tumors sequenced by MSK-IMPACT from January 2014 to October 2020. The tumor diagnoses were confirmed by a board-certified pathologist and categorized into 60 distinct tumor types based on morphology and site of primary tumor, according to Oncotree grouping (http://oncotree.mskcc.org, last accessed June 16, 2019), according to the available clinical data at the time of sequencing. All sequenced solid tumors that passed quality control, including confirmation of adequate genomic coverage and lack of contamination, were included as part of the study to identify viral sequences in the tumor tissue.
Virus Read Detection
The study was based on paired reads, including human-virus chimeric reads, which improve virus specificity and eliminate misallocation of reads between samples within individual runs. Samtools version 1.7 was used to extract paired unmapped reads from processed BAM files of MSK-IMPACT clinical samples into FASTA files.17 The unmapped reads from each sample were queried for viral content using blastn 2.9.0+ against all human viruses from the National Center for Biotechnology Information Virus database (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/#, last accessed August 10, 2020) using the following settings: strand both, word_size 28, evalue 1 e-10, and perc_identity 90.18 The blastn settings were tuned to improve specificity based on experiments designed to enhance sensitivity and specificity for a subset of cases known to be high-risk (HR) human papilloma virus (HPV) positive. Notably increasing the minimum word size identity greatly reduced candidate matches and enhanced specificity. KronaTools (v2.7) function ClassifyBLAST was used to annotate corresponding National Center for Biotechnology Information Taxonomy (https://www.ncbi.nlm.nih.gov/taxonomy, last accessed July 31, 2020) to the aligned hits using the GI identifier from blastn output.19 To account for the presence of the Epstein-Barr virus (EBV) genome in GRCh37, paired reads from the original processed BAM files aligned to NC_007605 were counted. No other viral genomes were incorporated in the reference genome utilized. TaxonKit 0.6.0 was used to categorize each virus taxonomic identifier within a specific species and genus.20 The python script for implementing this pipeline is provided as Supplemental Code S1.
High-Risk HPV Validation with ISH
The standard-of-care, clinically validated high-risk HPV in situ hybridization (ISH) test for evaluation for the presence of HR HPV in tumor tissue was used to test performance of the method. Tumors evaluated by the RNA HR HPV ISH and MSK-IMPACT testing were identified. The cases evaluated by HR HPV ISH were tested as a part of routine surgical pathology workflow. Methods for HPV ISH have been described previously.21 The probes were designed to detect the E6 and E7 genes of seven HR HPV genotypes (HPV-16, HPV-18, HPV-31, HPV-33, HPV-35, HPV-52, and HPV-58). The pathologist's interpretation of the HR HPV ISH was documented, and the slides for each case were obtained to verify the results.
HR HPV Validation with Isothermal Amplification
Performance characteristics were assessed in comparison to the AmpFire HPV Detection and AmpFire Genotyping HPV assay (Atila BioSystems Inc., Mountain View, CA), using an additional aliquot of extracted DNA. Twenty-eight cases from which ISH was performed had residual DNA for testing by isothermal amplification. The remaining cases were selected randomly within positive and negative classes from cases with residual DNA from MSK-IMPACT with a target prevalence of 50% based on MSK-IMPACT HPV results. This assay uses isothermal amplification real-time fluorescent detection that is validated to detect and classify 15 high-risk HPV types.22
Analysis of EBV-Associated Tumors
Epstein-Barr Early Region (EBER) in situ hybridization is commonly performed in clinical laboratories to identify the presence of EBV virus.23 A retrospective analysis of the tumors with EBV ISH was performed as part of the clinical workflow for tumors. For cases in which >70 reads of EBV were identified, additional EBV ISH tests were performed when tissue was available. EBV ISH was performed on a select group of tumors with <70 reads to identify any nontumor cells with identifiable virus.
Virus Associations with Tumor Type and Mutations
Viruses detected with two or more paired reads were associated with the respective Oncotree tumor type. The two-read cutoff is based on optimal threshold for detecting HPV based on above validation. The frequency of a virus type in a specific tumor type was compared with the frequency of the same virus in all other tumor types combined using an odds ratio (OR). An instance was generated in CBioportal (https://www.cbioportal.org, last accessed September 28, 2020) that associated the tumor cases with the virus species and number of viral reads identified. The Groups feature was utilized to generate cohorts of virus-positive and tumor type–matched virus-negative tumors to determine enrichment or depletion of specific genomic event in virus-positive cases, reported as ORs. A validation cohort of 7814 consecutive clinical tumor cases not overlapping with the discovery cohort was identified. The validation cohort included samples analyzed by an updated version of the MSK-IMPACT panel in which probes for the human DNA viruses were added. Thus, the virus-positive tumors were enriched for virus reads and were not off-target captured reads as in the discovery cohort. Read threshold was calibrated for updated methods. The validation cohort was used to demonstrate that the novel findings of prior analyses can be replicated on an independent cohort using a different hybridization capture protocol.
Viral Integration Detection
A virus-human combined reference genome was generated utilizing the standard human reference (GRCh37) and adding 197 relevant viral genomes as additional contigs. Samples with ≥10 viral reads identified by the previously discussed viral identification pipeline were selected to identify the integration site. The complete complement of FASTQ files generated from sequencing was aligned to the virus-human reference. DELLY version 0.7.7, the structural variant detection algorithm currently employed in MSK-IMPACT pipeline, was used for identification of integration events.15 A translocation event is called by the algorithm when one end of the split read aligns to the human genome and the other to one of the virus genomes. This is consistent with an integration event, and the integration site is documented. In parallel, a previously reported integration event algorithm called GENE-IS version 1.0 was used, which has been used as a viral vector detection algorithm in gene therapy studies to validate the results from DELLY algorithm.24
Statistical Analysis
Statistical calculations were performed using R version 3.5.1. Receiver operating characteristic curve calculations were performed using the R package pROC, and CIs were calculated using bootstrapping methods.25 ORs were calculated with R package fmsb with a 2 × 2 matrix, where the odds of the tumor type being positive for a virus were compared with the odds that the remaining cohort was positive for the same virus. Each analysis was performed independently. Haldane-Anscombe correction was applied as appropriate. Bonferroni adjustment of the P value was utilized for all analyses where adjustment for multiple hypotheses was needed. Comparison of tumor mutation burden in virus-positive tumor was performed using unpaired t-test on the normally distributed log10 of sample-level tumor mutation burden. The R script for statistical analysis is provided (Supplemental Code S2).
Results
Detection of Viral DNA in Routine Clinical Genomic Testing
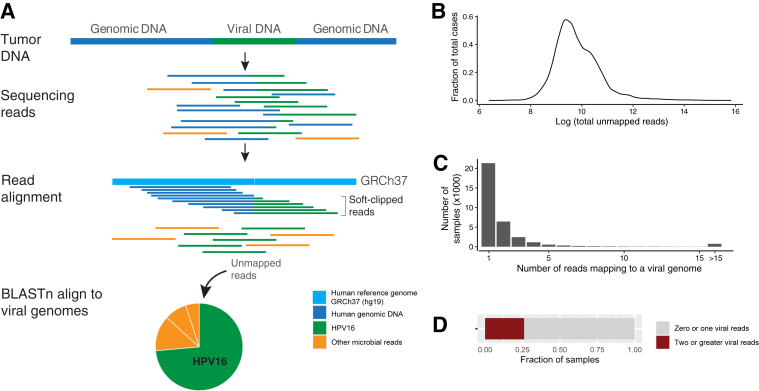
Hybridization-capture–based NGS assays retain a small amount of off-target genomic DNA and the subsequently sequenced nonhuman DNA. Because of the high read depth (500× to 1000×) generated through targeted clinical sequencing applications, these reads occur at a frequency that the analysis of the nontarget reads can be exploited to identify clinically relevant viruses (Figure 1A). Sequencing data from 48,148 samples (42,846 patients) analyzed by MSK-IMPACT, the clinical NGS assay developed at Memorial Sloan Kettering Cancer Center, were used to identify, on average, 12,096,946 unique paired sequencing reads per sample. On average, 32,810 paired reads (median, 15,791; range, 595 to 7,421,821; 0.3% of all paired reads) did not map to the human genome (Figure 1B), and 15 (median, 1; range, 0 to 78,998) paired human virus reads were identified per sample. Just 26.1% of cases had >1 human virus read (Figure 1, C and D).
Figure 1.
Summary of virus discovery method. A: DNA sequencing reads derived from tumor-associated microbes that do not align to the human genome. Both reads that are chimeric (ie, human plus virus) and reads of only virus were analyzed for the presence of specific viruses. Tumor-associated bacteria and fungal reads were also detected but are not part of this analysis. B: Density plot of all unmapped reads across the cohort. C: Distribution of number of reads that align with viral genomes in a given sample. D: Rate of detection of greater than one read of virus in a given sample. HPV, human papilloma virus.
Comparison of Virus Detection Method with Reference Assays
To determine a threshold above which a sample would be considered to harbor a virus, two well-characterized viruses with existing clinically validated orthogonal tests, HR HPV and human herpes virus (HHV) 4/EBV, were chosen. The standard-of-care method for detection of HR HPV in formalin-fixed, paraffin-embedded (FFPE) specimens is RNA in situ hybridization.21 A total of 122 specimens, enriched for cervical and head and neck cancer, had both ISH and MSK-IMPACT testing performed as part of clinical testing; and concordance between the two methods was 96% (90% sensitivity, and 100% specificity). To assess the performance of the MSK-IMPACT virus detection method with unbiased sample collection, 108 samples were tested with AmpFire Genotyping HPV, an isothermal amplification assay that is a highly sensitive platform for detecting HR HPV DNA in FFPE material.22 The complete list of validation samples and results are provided (Supplemental Table S1). Of the 122 samples with ISH, 28 had residual DNA available from sequencing available for testing by isothermal amplification.
Receiver Operating Characteristic Curve Analysis
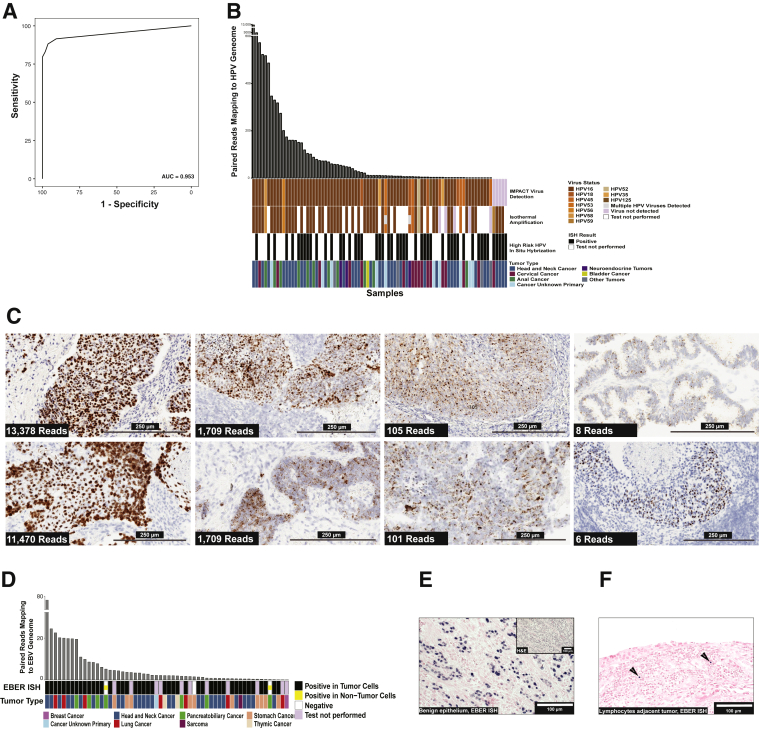
Receiver operating characteristic analysis comparing NGS read analysis for HR HPV with isothermal amplification showed an area under the curve of 95.3% (95% CI, 91.3%–99.2%, by DeLong bootstrapping method) (Figure 2A). The optimal threshold for determining presence of HR HPV was the presence of two or more paired virus reads. At the two paired read threshold, the specificity of the nonmapping paired read analysis from MSK-IMPACT is 94.3% (95% CI, 88.7%–100%) and sensitivity is 92.7% (95% CI, 78.2%–98.2%). Figure 2B summarizes the results of validation samples that were positive for HR HPV by one of the three methods, including demonstration of 100% concordance in HPV genotyping between methods when a virus was detected by both isothermal amplification and MSK-IMPACT. Supplemental Table S1 includes the results from all validation cases, including negative samples. The performance characteristics for virus detection were independent of total sample reads and total nonmapping reads.
Figure 2.
Validation of virus detection using nonmapping reads. A: Receiver operating characteristic analysis for high-risk (HR) human papilloma virus (HPV) detection with isothermal amplification detection method as the standard. Area under the curve was 0.953, and the optimal threshold for identifying HR HPV was two or more reads. B: Comparison of Memorial Sloan Kettering–Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) analysis, isothermal amplification, and in situ hybridization (ISH) results for detection of HR HPV. The figure displays cases where virus was detected by any of the three methods. Each vertical bar represents a single case. The bar plot at top shows the number of paired reads of the virus detected by MSK-IMPACT. The HPV genotype is shown by colors to compare the genotype detection between IMPACT and isothermal amplification. The bottom row shows the tumor type for each case as indicated in the legend. C: Demonstration of increased HR HPV RNA expression (brown signal) in tumor cells with a large dynamic range of HR HPV reads detected by MSK-IMPACT. The number of reads correlates with a combination of the intensity of RNA expression by ISH in tumor cells, the proportion of tumor cells where virus RNA expression was detected, and the proportion of neoplastic cells on the slide. Low tumor proportion is frequently a factor in cases with lower virus reads. D: Comparison of Epstein-Barr Early Region (EBER) ISH and IMPACT for cases with >70 EBV reads detected by next-generation sequencing (NGS) read analysis and the respective tumor types. E: EBER ISH of a case with >70 EBV reads where virus was localized to benign epithelium (blue signal indicates positive cells). F: EBER ISH of tumor tissue with <70 EBER reads by NGS read analysis where virus localized to tumor-associated lymphocytes (arrows point to positive cells). Scale bars: 250 μm (C); 100 μm (E and F). AUROC, area under the receiver operating characteristic curve; H&E, hematoxylin and eosin.
Assessment of Discrepant Cases and Quantitative Nature of Paired Virus Read Detection
The study explored cases that were discrepant among methods. Figure 2C shows the comparative signal by RNA ISH in samples with a broad range of virus reads. As the total number of paired reads detected decreased, the strength of ISH signal also decreased, which suggests the viral reads detected correlate with the level of RNA expression and thus the amount of virus nucleic acid in samples. The relationship between the strength of staining by ISH and the total number of reads detected by NGS suggested that analysis of nonmapping reads is semiquantitative. The other source of discrepancy between NGS and reference methods was cases with low tumor content relative to surrounding inflammatory cells, which is a known source of false-negative mutation calls for somatic mutation detection by NGS.26 The discrepant cases among the three methods, including the discrepancy between ISH and isothermal amplification, occurred when the total virus reads from MSK-IMPACT were <6 paired virus reads. These cases also had weak signal for virus by ISH or had low tumor content. This finding suggests that samples with low reads detected by NGS are more challenging cases to detect the virus by all methods, presumably because these samples have a lower level of viral nucleic acid. Differences between various reference methods for virus detection are well described.27 The discrepancies have been ascribed to technical limitations of different assays, but these results suggest that sample dependencies, such as lower levels of virus, are also a factor. Overall, the validation experiments suggest that the analysis of nonmapping NGS reads has similar performance to reference methods for detecting HR HPV.
Assessment of Detection of EBV
An EBV RNA ISH test is the standard-of-care test for EBV in solid tumors. The study identified 39 cases from the MSK-IMPACT cohort with EBV ISH testing performed during routine pathology assessment. The ISH testing was enriched in tumors with a morphologic phenotype, called lymphoepithelial-like carcinoma (LE-LC) that is associated with EBV positive epithelial tumors, which was observed in nasopharyngeal, esophagogastric, thymic, and small bowel/ampullary tumors. The study found cells with EBV signal by RNA ISH in all samples that contained NGS reads and for which EBV ISH was performed clinically. However, EBV presents a unique challenge not seen in other viruses. EBV not only causes oncogenic transformation but also is a common chronic infection that can be reactivated when patients experience immunosuppression, such as during chemotherapy.28 Thus, EBV is the most common virus observed in the data set, with 22% of samples having two or more EBV reads. Comparison of samples with EBV confirmed by ISH was used to establish that all cases with EBV expression in the tumor cells contained ≥70 reads by NGS. A total of 90 cases within the cohort had ≥70 EBV reads. A total of 47 of these cases had available tissue for EBV ISH to be performed, including those with previous clinical testing. The results are summarized in Figure 2D. One case of gastric cancer with LE-LC morphology was negative for ISH but had >2000 reads detected by NGS, suggesting the RNA ISH was a false negative. Two cases with >70 EBV reads were negative in the tumor cells, but the EBV ISH was positive in the benign epithelium adjacent to the tumor and not within the tumor cells (Figure 2E). This demonstrates a clear source of virus presence when the virus is unrelated to the malignancy. This situation can be resolved in routine clinical practice by EBV ISH. EBV infection of benign epithelium has been previously described but not in the setting of adjacent malignancy.29 The source of EBV signal below the 70-read threshold was reactivation of EBV in tumor-associated lymphocytes (Figure 2F).
Confirmation of Expected Tumor-Virus Associations
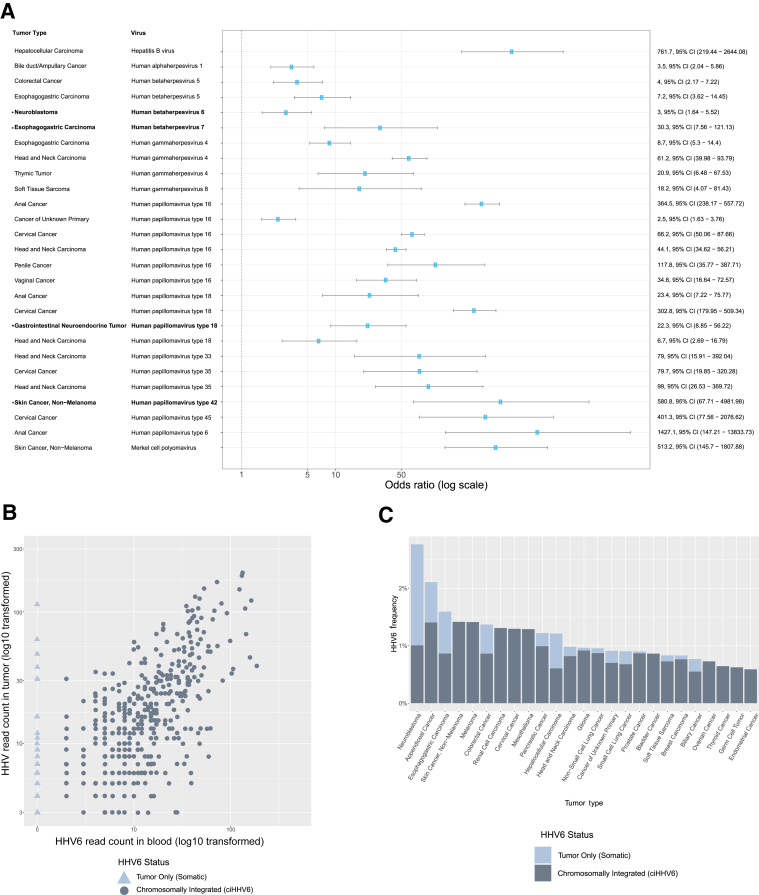
The viral read analysis was extended to a larger cohort of 48,148 samples sequenced prospectively between January 2014 and September 2020. Considering taxonomies observed in two or more samples, a total of 82 unique virus taxonomies from 51 distinct species of virus were present in 12,566 clinical tumor samples. The frequency of virus-tumor type combinations was compared with frequency of the same virus in the full cohort of all other tumor types. Each analysis was performed independently as the presence of one virus does not exclude the presence of another. Figure 3A shows the tumor-virus pairs with four or more instances in the cohort and for which the pair frequency is significantly increased over controls (lower bound 95% CI > 1 and significant P value after Bonferroni adjustment for each taxonomy tested per tumor type). This analysis was used to recapitulate known virus-tumor type relationships: various HPV genotypes in cervical, anal, and head and neck cancer; EBV in LE-LC head and neck, thymic, and esophagogastric cancer; Merkel cell polyoma virus in nonmelanoma skin cancer (ie, Merkel cell carcinoma); HHV8 in sarcomas (ie, Kaposi sarcoma); and hepatitis B in hepatocellular carcinoma.
Figure 3.
Association between viruses and tumor types in 48,148 tumors. A: Forest plot demonstrating the virus-tumor type relationships that are enriched with statistical significance (P < 0.005), the cutoff was determined for Bonferroni adjustment given multiple hypotheses for each tumor type. Error bars show the 95% CI. Associations are arranged by virus. Associations in bold and labeled with asterisk are novel associations. B: Scatterplot showing the virus reads of human herpes virus 6 (HHV6) in tumor (y axis) compared with peripheral blood (x axis). The light blue triangles indicate samples that have virus detected only in tumor but not in blood. Both axes are log10 transformed. C: Bar chart showing the frequency of HHV6 detected in different tumor types and proportion of cases that are chromosomally integrated (ciHHV6) versus tumor only (somatic). Neuroblastoma is the only tumor type that is statistically enriched for HHV6, and the enrichment is from samples with HHV6 in tumor-only samples.
Viruses Associated with Organ-Specific Infections
Viruses that are common infections in the primary organ of the tumor type but are not likely to be associated with the development of malignancy are also observed: human herpes virus 5/cytomegalovirus in colorectal and esophagogastric cancers; and herpes simplex virus (human alphaherpesvirus 1) in esophagogastric carcinoma.30, 31, 32 Although several cases of herpes simplex virus were observed in esophagogastric cancer with high read counts, herpes simplex was statistically enriched only in bile duct/ampullary cancers. It is possible that these opportunistic infections may be related to immunosuppression related to cancer therapy or to a permissive immune system that contributed to the development of malignancy.33,34
Novel Virus-Tumor Associations
An association between HHV7 and esophagogastric carcinoma has not previously been observed. HHV7 is also observed in salivary gland cancers, but there are only two examples. Studies have shown varying rates or persistent shedding of HHV7 in saliva of patients, but no association between esophagogastric tumors and HHV7 has been described.35 The enrichment of HPV18 in a subset of gastrointestinal cancer with neuroendocrine morphology is a novel observation. The HPV18-positive neuroendocrine tumors tend to be high-grade tumors and involve the distal colon. The exclusive presence of HPV42 in a rare form of nonmelanoma skin cancer called digital papillary adenocarcinoma is also not described in the literature, although there has been speculation that the entity is associated with HR HPV.36 HPV42 does not belong to the group of HR HPV and thus targeted PCR assays and ISH for HR HPV would not detect the virus.
HHV6 Enrichment in Neuroblastoma
HHV6 is not an oncogenic virus and is typically associated with childhood illness and transient infection associated with immunosuppression. HHV6 is also integrated into the germline [chromosomally integrated HHV6 (ciHHV6)] and inherited in 1% to 2% of the population. ciHHV6 exhibits mendelian inheritance.37,38 There is no compelling evidence that ciHHV6 predisposes individuals to malignancy. As such, the expected proportion of cases positive for this virus should be equivalent across tumor types. However, enrichment of the virus was observed in neuroblastoma. MSK-IMPACT utilizes a paired peripheral blood sample for the purpose of identifying and excluding germline polymorphisms as somatic events in tumors. When ciHHV6 is present, we expect the virus to be observed in both the tumor and the paired peripheral blood sample. Within the cohort, 702 (1.4%) tumor samples had detectable HHV6. Figure 3B shows the quantity of reads in HHV6-positive tumors and the corresponding number of reads in the paired blood sample. The triangles in the figure represent the rare cases where HHV6 is present in the tumor but not in the blood, which suggests the virus is not chromosomally integrated and thus in tumor only. For all other viruses detected in tumor, the corresponding virus reads were rarely detected in the blood, and when present, they were suggestive of tumor cell-free DNA with integrated virus or a systemic infection, such as EBV infection. For HHV6, 83% of virus-positive tumor samples also had virus reads detected in the blood sample. On the basis of this, these cases were presumed to be ciHHV6. However, 17% of HHV6-positive samples were found to have HHV6 only in the tumor, which appears to represent a different biological case than ciHHV6.
Tumor-Only HHV6 Enriched in Neuroblastoma
Whether different tumor types had consistent levels ciHHV6 versus tumor-only HHV6 was investigated. Figure 3C shows the relative frequency of ciHHV6 in the tumor types. As expected, based on population frequency of ciHHV6, most tumor types showed a frequency of HHV6 at approximately 1%. Neuroblastoma was the only tumor type statistically enriched for HHV6 and had HHV6 positivity 3.0 times the average. Interestingly, for neuroblastoma, the increased frequency of HHV6 is fully attributable to HHV6-positive tumors with no virus detected in blood. The OR for cases with HHV6 only detected in tumor for neuroblastoma was 13.7 (95% CI, 6.3–30.0; P = 2.2 × 10−16). This makes the relationship between neuroblastoma and HHV6 compelling and worthy of more detailed investigation.
Validation Cohort
The validation cohort contained 7813 consecutive cases from the clinical testing cohort with a similar variety of different tumors. These cases were unique and independent from the discovery cohort. The four novel virus tumor type associations were the focus of this analysis. The associations for which there were statistical power on the validation cohort are available in Supplemental Figure S1A. HHV6 was enriched in neuroblastoma, colorectal, and esophagogastric cancer in the validation cohort. This validation cohort contained 108 neuroblastoma cases, and 4 of the cases are positive for HHV6. All colorectal and esophagogastric tumors positive for HHV6 were also positive in the blood, consistent with ciHHV6. All neuroblastoma cases with HHV6 reads in the tumor had zero HHV6 reads in the blood, consistent with tumor-specific HHV6. As in the discovery cohort, neuroblastoma was significantly enriched for tumor-specific HHV6 (OR, 13.7; 95% CI, 4.6–41.1; P = 8.7 × 10−10) in the validation cohort. HHV7 was enriched in esophagogastric cancer and salivary gland, as in the discovery cohort. One case of digital papillary carcinoma was present in the validation set that contained 800 reads of HPV42. Images of the histology and RNA ISH are provided in the Supplemental Figure S1B. The validation cohort did not confirm the association between HPV18 and gastrointestinal neuroendocrine tumors. The discovery cohort showed the HPV18-positive tumors were high-grade gastrointestinal neuroendocrine tumor. The validation cohort contained four high high-grade gastrointestinal neuroendocrine tumors, and zero were positive for HPV18. Two of the four high-grade gastrointestinal neuroendocrine tumor samples were positive for other HPV genotypes in the validation cohort.
Oncogenic Virus Status and Genomic Alterations
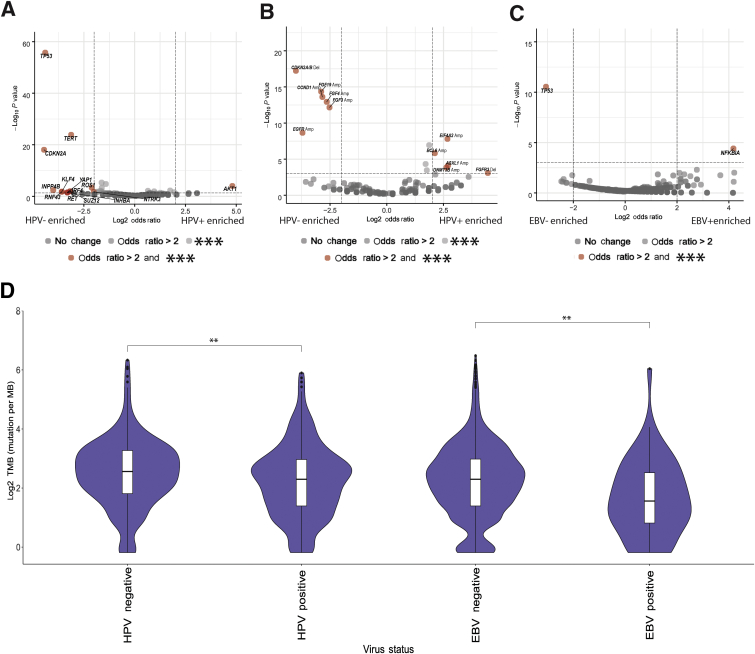
A total of 538 patients with HR HPV-positive tumors were compared with a tumor type–matched control cohort consisting of 643 presumed HR HPV-negative head and neck, cervical, anal, vaginal, and penile squamous cell carcinoma cases. HR HPV-positive cases showed a decreased frequency of mutations in TP53 (P = 1.9 × 10−79), TERT promoter (P = 1.2 × 10−31), and CDKN2A (P = 4.9 × 10−18) compared with HR HPV controls. Inversely, HR HPV-positive tumors were enriched for AKT1 (P = 1.6 × 10−4) mutations (Figure 4A). Specific copy number alterations were also differentially distributed in the two cohorts. Chromosome region 11q13.3 (P < 1.0 × 10−12) and EGFR (P = 2.2 × 10−9) amplifications as well as 9p21 deletions (P < 5.3 × 10−15) were enriched in HPV-negative tumors. The genes BCL6 (P = 1.1 × 10−7), EIF4A2 (P = 1.6 × 10−8), DNMT3B (P = 1.4 × 10−4), and ASXL1 (P = 7.6 × 10−5) were more frequently amplified, and FGFR3 (P = 6.9 × 10−13) was more frequently deleted, in HR HPV-positive tumors (Figure 4B).
Figure 4.
Genomic associations with high-risk (HR) human papilloma virus (HPV) and Epstein-Barr virus (EBV). A–C: Volcano plots showing oncogenic virus-positive tumors. A–C: The dashed lines representing predetermined thresholds for displaying events enriched, with vertical and horizontal dashed lines representing an odds ratio of ±2 and P value of −log10(0.001), respectively: HR HPV mutations (A), HR HPV copy number alterations (B), and EBV mutations (C) are enriched (right side of plots) or depleted (left side of plot) in virus-positive tumors. The x axis is negative log10 of P values. Gene names are displayed for genes that are enriched/depleted in virus-positive tumors with P < 0.001. The P-value threshold is Bonferroni-adjusted for the multiple genes tested. D: Tumor mutation burden (TMB) comparison between virus-positive tumors and tumor type–matched negative controls. Statistical analysis is via an unpaired t-test. ∗∗P < 0.01 (D), ∗∗∗P < 0.001 (A–C). Amp, amplification; Del, deletion.
EBV-Positive Tumor Associations
The analysis of EBV-positive tumors was restricted to the tumors with >70 EBV reads. A total of 90 tumors were compared with tumor type–matched controls. EBV-positive tumors were enriched for mutations in NFKBIA (P = 3.9 × 10−5) (Figure 4C), compared with negative controls, whereas EBV-negative controls showed increased frequency of TP53 (2.9 × 10−11). TERT promoter mutation (P = 0.04) was also enriched in EBV-negative tumors, but the result was not significant after multiple hypotheses correction. The copy number alterations showed no significant differences between EBV-positive tumors and EBV-negative controls.
Tumor Mutation Burden and Oncogenic Viruses
Both HPV- and EBV-associated tumors had a significantly lower tumor mutation burden (P = 0.003 and P = 0.003, respectively) than tumor type–matched controls (Figure 4D). The comparatively low tumor mutation burden in virus-associated tumors may be related to the expression of viral proteins that serve similar functions to activated oncogenes and/or inactivated tumor suppressor genes in virus-negative tumors. For example, lower rates of TP53 and TERT mutations were observed in virus-positive tumors compared with virus-negative tumors.
Virus Integration
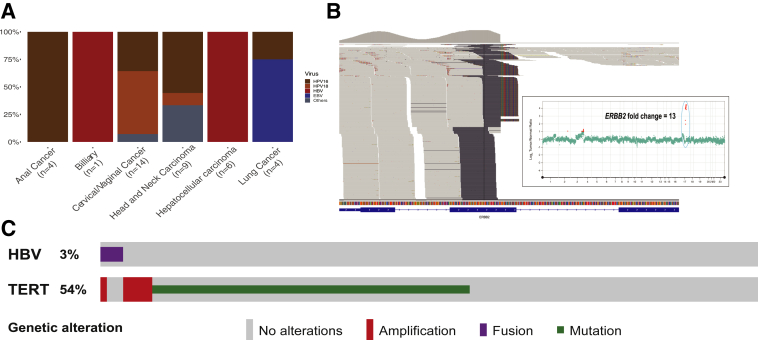
Viral integration sites within the human genome were observed by a modified analysis pipeline. A total of 619 samples with ≥10 viral reads were selected for viral integration analysis. A total of 18% of the samples tested showed evidence of integration with two or more chimeric reads containing both virus and human genome detected by at least one algorithm (Figure 5A). HPV16 and HPV18 were the most common viruses with detected integration sites in anal, cervical/vaginal cancers, and head and neck squamous cell carcinomas. Interestingly, an associated copy number change at the human genome integrations site was observed. Figure 5B shows ERBB2 amplification directly associated with integration site of HPV16 at the ERBB2 locus. This suggests that viral integration may drive oncogenesis not only by the expression of viral genes, as has been extensively described, but also by instigating genomic amplification of human oncogenes. Genomic amplification associated with virus integration has been proposed as a mechanism of oncogenesis in tumors based on tumor cell line evaluation.39 However, the authors are not aware of a clear demonstration of such an event in a clinical tumor sample. In the data set, HBV was recurrently integrated in the TERT promoter in hepatocellular carcinomas, which has been previously described.14,40 HBV integration in the TERT promoter region was mutually exclusive with TERT promoter mutations in our data set (Figure 5C).
Figure 5.
Analysis of virus integration identified by MSK-IMPACT. A: Bar plot showing the distribution of tumor types and viruses in which viral integration was detected. B: Genome browser view showing integration of human papilloma virus 16 (HPV16) within ERBB2 gene. The internal panel shows the whole genome log-ratio plot for the tumor, demonstrating 13-fold amplification of ERBB2 associated with the HPV16 integration, highlighted by the green circle. C: Oncoprint of hepatocellular carcinoma cases from cohort, demonstrating mutual exclusivity between hepatitis B virus integration and TERT mutations. EBV, Epstein-Barr virus.
Clinical Applications
In the course of this study, several virus-associated tumors were discovered at the time of sequencing, which would not have been identified without the current analysis. This was most commonly observed in EBV-positive tumors less commonly associated with EBV, such as LE-LC of the thymus, lung, and pancreatobiliary cancers. In several of these cases, especially in lung cancer, the lymphoepithelial-like phenotype was not fully evident. The observation of morphologically occult LE-LC of the lung along with reports of this entity41 has led the pulmonary pathologists to implement EBV ISH testing on all never smoker patients with squamous cell lung cancer. An EBV-positive cholangiocarcinoma case was discovered and confirmed by ISH. Protocols using immunotherapy strategies to target the EBV-specific antigens are currently being undertaken when occult EBV tumors are identified. Thus, detecting EBV as part of clinical genomic sequencing will be valuable with tumors not expected to be EBV-positive based on routine pathologic evaluation.
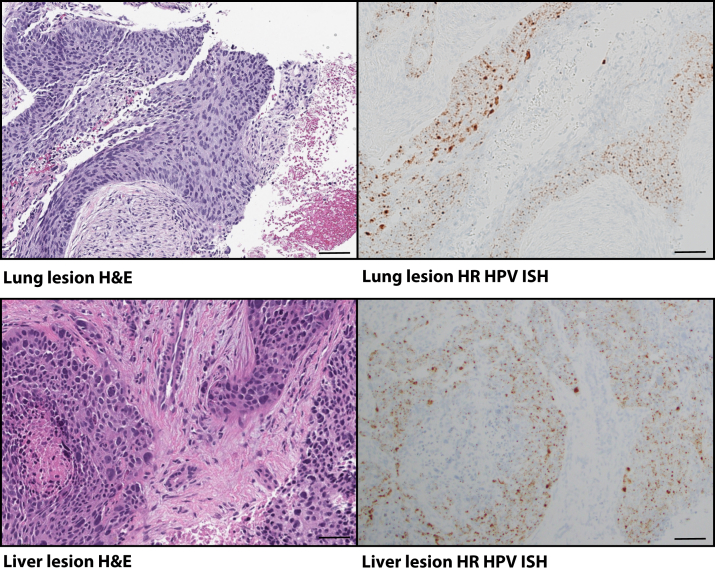
An impactful demonstration of the clinical utility of comprehensive virus detection for the management of cancer patients was that of a 55-year–old woman with a documented smoking history. The patient was found to have a lung mass. The lung mass was biopsied, and histologic examination was consistent with squamous cell carcinoma. The patient was diagnosed with lung squamous cell carcinoma and was managed with standard therapy for lung cancer. Subsequently, the patient developed a liver metastasis. The metastatic lesion was confirmed by histology and was then sent for molecular testing. HPV16 reads were identified at the time of sequencing. HR HPV ISH confirmed the presence of virus on both the lung and the liver biopsy (Figure 6). The patient had a remote history of early-stage cervical cancer that was believed to have been cured by simple excision. Knowing the HPV16 status, the diagnosis was revised to metastatic cervical cancer. This vignette demonstrates the potential for prospective and routine virus analysis to inform cancer therapy.
Figure 6.
High-risk (HR) human papilloma virus (HPV) in unexpected metastatic tumor. Photomicrographs of the squamous cell carcinoma recharacterized to be metastatic cervical cancer. Left panels: Hematoxylin and eosin (H&E)–stained tumors. Right panels: Corresponding HR HPV in situ hybridization (ISH) in subsequent sections showing strong signal for the virus in tumor cells in both lesions from the lung and liver. Scale bars = 100 μm.
Discussion
The study described here is the largest and most comprehensive study of human DNA virus detection in cancer reported in the literature and the first clinical validation and determination of performance characteristics of metagenomics techniques by analyzing nonhuman reads from cancer tissue. The investigation includes 48,148 individual tumor samples of diverse tumor types prospectively sequenced in a clinical setting. This analysis demonstrates the feasibility of detecting (and potentially reporting) tumor virus status for use in routine clinical management of cancer patients. Obtaining viral status in tissue has historically been a major challenge because of the cost and technical hurdles not addressed by other testing methods.27 Virus discovery in clinical practice by molecular methods has been hindered by the inherent challenge of traditional molecular methods on tumor specimens, which are routinely processed as FFPE tissue for pathologic examination. Methods currently employed for such analyses for HR HPV experience various challenges related to sensitivity and specificity due to nucleic acid degradation in FFPE tissue. The most common method for detecting viruses in FFPE tissue, in situ hybridization, lacks ability to genotype the viruses.27 These methods also do not allow for the detection of multiple different virus classes in a single test. Because of these challenges, testing for the presence of multiple viruses in tumor tissue is not routinely performed, and testing for single viruses is limited to narrow clinical scenarios. Establishing a technique with comparable sensitivity and specificity to legacy molecular methods that allows for the detection of the common human DNA viruses would be of tremendous value for the management of cancer patients. The technique described herein utilizes data previously acquired for clinical tumor sequencing. Because this is a bioinformatic technique, it requires no additional sequencing, and virus detection can be achieved with minimal additional cost. This technique paves the way for clinical reporting of viral status of tumors along with the somatic mutation report and offers the potential to add significant value to the clinical genomic sequencing of tumor tissue.
The value of virus identification in routine clinical practice, specifically when performed in tandem with somatic mutation detection, has many appealing aspects for patient management and clinical investigation. Aside from providing insights into etiology, virus status can be utilized to resolve diagnostic challenges, such as when a metastatic lesion does not have a clinically evident primary source (cancer of unknown primary). It can also be prognostically significant and can function as a predictive biomarker for treatment response (radiotherapy and immunotherapy).42,43
This study validates the accuracy of the digital subtraction method on DNA reads that do not align to the human genome from a large-panel NGS assay without virus-specific probes. By cross validating the results with multiple methods across tumor types and with different virus species, the method is demonstrated to be readily applicable to data from a clinical hybridization capture NGS sequencing assay. The validation data allow for extension of the analysis to discover novel tumor-virus associations.
The novel tumor-virus associations of HHV6 in neuroblastoma and HHV7 in esophagogastric cancer were validated using an independent data set that specifically enriches the sequenced reads for viral reads. To the best of our knowledge, the association between neuroblastoma and HHV6 is novel. To rule out adrenal gland HHV6 colonization as a cause, 297 non-neuroblastoma adrenal gland samples from the cohort were observed. A total of 112 non-neuroblastoma adrenal gland primary tumors and 185 tumors metastatic to the adrenal gland were present in the cohort and were all negative for HHV6. Furthermore, the cohort contained two patients with neuroblastoma with both primary and distant metastasis sequenced. In both cases, the primary and metastatic tumor samples were positive for HHV6, and the peripheral blood samples were negative, ruling out systemic infection and isolating the virus to the tumor tissue in these patients. The study does not allow for conclusive evidence of a causal relationship between HHV6 and neuroblastoma but shows a strong and unique relationship not observed in other tumor types. Additional investigation is warranted to better understand the relationship between HHV6 and neuroblastoma.
The final novel association is the relationship of HPV42 to digital papillary carcinoma. The pathology department's clinical in situ hybridization laboratory has validated an HPV42 ISH assay that is now used routinely in identifying digital papillary adenocarcinoma at our institution. All samples to date where HPV42 reads were found in the tumor have been confirmed with the ISH assay, and zero samples with this histology have lacked the virus. A technique of unbiased detection of virus DNA allows for the opportunity to investigate not only the role of known viruses in oncogenesis, but also potentially undiscovered viruses by using established and emerging metagenomic virus discovery techniques.44,45 Having access to these data for discovery by data analysis alone allows for resources to be dedicated to investigating the role that viruses might play in oncogenesis and considering virus-informed therapies rather than performing laborious single virus discovery assays.
This analytic approach also allows for defining the genomic landscape of tumors that contain virus compared with tumor type–matched controls. With virus data collected in tandem with genomic data, more extensive analyses can be performed to better understand the interactions of the tumor genome with oncogenic viruses. The largest previous study of viruses in tumors was the Pan-Cancer Analysis of Whole Genomes consortium study utilizing whole genome and transcriptome sequencing from 2658 The Cancer Genome Atlas tumor samples.14 Whole transcriptome sequencing has some benefits for detecting a greater number of viral reads and more chimeric reads, and the ability to detect RNA viruses. The findings of the Pan-Cancer Analysis of Whole Genomes study and the current study are largely complementary. The strength of the Pan-Cancer Analysis of Whole Genomes study was the ability to detect large numbers of viral integration sites in virus-positive tumors from RNA sequencing compared with the relatively small number of integration events detected using DNA reads from targeted clinical sequencing. The depletion of TP53, CDKN2A, and TERT mutations in HPV-positive tumors was noted in both studies. The current study, however, being an order of magnitude larger, allows for increased statistical power to discover novel virus-tumor type associations and some novel virus-mutation associations. The current study also has the advantage of being able to validate the bioinformatics method against clinical laboratory tests to determine the threshold for detecting positive cases and determining sensitivity and specificity for virus detection in a clinical setting.
Although the sequencing is optimized for capturing specific regions of the tumor genome and the viral reads are a convenient byproduct of hybridization capture technology, the sensitivity and specificity observed herein are remarkable. Sensitivity can be improved by optimization of both the sequencing assay and bioinformatics analysis. In the updated MSK-IMPACT clinical assay used as the validation cohort in this study, hybridization capture probes were designed and implemented to enrich for DNA virus-specific sequences. Preliminary validations confirmed that improved sensitivity for the viruses and integration site detection can be achieved with this strategy. As an example, the method with virus-specific probes identified at least 10-fold greater reads per case on average in HPV16-positive cases compared with the original method.
Although based on the HR HPV assessment the technical specificity is high, viruses can be present in tumor tissue even when the virus is not the causal agent. For example, patients can have viral infections in nonneoplastic cells near tumor or in tumor-associated lymphocytes. Therefore, caution must be considered in interpretation of these data, and direct visualization techniques or virus integration analysis should be performed to confirm the role of the virus in the cancer.
Multiple molecular techniques are currently employed in clinical practice to detect various oncogenic viruses.46, 47, 48 However, historical methods only allow for the detection of a single virus type per test, and each test requires additional time, cost, and tissue, which has made testing of tumors for viruses uncommon in routine management of cancer patients. As hybridization capture NGS becomes more common in clinical oncology, a virus detection mechanism incorporated with somatic mutation detection offers the opportunity to augment clinical practice for cancer patients. The emergence of genomic medicine and targeted molecular therapy now allows for the efficient detection of mutations in many genes in thousands of cancer cases.16 A similar trajectory may be possible for virus-relevant cancer therapy as more advanced techniques for virus discovery are implemented with advances in virus antigen-directed immunotherapy.49 This analysis of nonmapping reads in clinical sequencing data offers the highest potential to understand tumor-virus relationships across cancer tumor types. The technique has the potential for high utility both in clinical practice and for understanding the roles of viruses in cancer.
Footnotes
Supported in part through the NIH/National Cancer Institute Cancer Center Support grant P30 CA008748 and supported by the Marie-Josée and Henry R. Kravis Center for Molecular Oncology.
Disclosures: X.C. and Y.W. are employees of Atila Biosystems Inc., which has developed a commercial assay for the detection of tumor-associated viruses.
Current address of S.M., Daichi Sankyo, Tokyo, Japan; and of Y.-W.T., Cepheid, Sunnyvale, CA.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2022.01.011.
Author Contribution
C.M.V. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Supplemental Data
Virus tumor associations in validation cohort. A: Forest plot demonstrating the statistically significant (P < 0.005) virus-tumor type relationships that are enriched in the validation cohort of 7813 consecutive samples using the updated version of the assay with virus-specific probes. Error bars indicate the 95% confidence interval. Associations are arranged by virus. B: Photomicrogaphs of a case of digital papillary adenocarcinoma (DPA). Scale bar = 100 micrometers. The top panel indicates H&E and the bottom panel indicates an HPV ISH using RNAScope probes specific to the HPV42 RNA, designed internally to assist with clinical identification of DPA based on the association established by this study.
Microbiome Extraction Pipeline
Stastical Analysis
References
- 1.Rous A. Sarcoma of the fowl transmissible by an agent separable from the tumor cells. Am J Med Sci. 1911;142:312. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.zur Hausen H., Schulte-Holthausen H., Klein G., Henle W., Henle G., Clifford P., Santesson L. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature. 1970;228:1056–1058. doi: 10.1038/2281056a0. [DOI] [PubMed] [Google Scholar]
- 3.zur Hausen H. Human papillomaviruses and their possible role in squamous cell carcinomas. Curr Top Microbiol Immunol. 1977;78:1–30. doi: 10.1007/978-3-642-66800-5_1. [DOI] [PubMed] [Google Scholar]
- 4.zur Hausen H. Papillomaviruses in anogenital cancer as a model to understand the role of viruses in human cancers. Cancer Res. 1989;49:4677–4681. [PubMed] [Google Scholar]
- 5.Dürst M., Gissmann L., Ikenberg H., zur Hausen H. A papillomavirus DNA from a cervical carcinoma and its prevalence in cancer biopsy samples from different geographic regions. Proc Natl Acad Sci U S A. 1983;80:3812–3815. doi: 10.1073/pnas.80.12.3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beasley R.P., Lin C.-C., Hwang L.-Y., Chien C.-S. Hepatocellular carcinoma and hepatitis B virus. Lancet. 1981;318:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 7.International Agency for Research on Cancer Working Group on the Evaluation of Carcinogenic Risks to Humans . World Health Organization; Geneva, Switzerland: 1997. IARC Monographs on the Evaluation of Carcinogenic Risk in Humans. [Google Scholar]
- 8.Saito I., Miyamura T., Ohbayashi A., Harada H., Katayama T., Kikuchi S., Watanabe Y., Koi S., Onji M., Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci U S A. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganem D., Varmus H.E. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 10.Feng H., Shuda M., Chang Y., Moore P.S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 2008;319:1096–1100. doi: 10.1126/science.1152586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cantalupo P.G., Katz J.P., Pipas J.M. Viral sequences in human cancer. Virology. 2018;513:208–216. doi: 10.1016/j.virol.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burk R.D., Chen Z., Saller C., Tarvin K., Carvalho A.L., Scapulatempo-Neto C., et al. Integrated genomic and molecular characterization of cervical cancer. Nature. 2017;543:378–384. doi: 10.1038/nature21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lawrence M.S., Sougnez C., Lichtenstein L., Cibulskis K., Lander E., Gabriel S.B., et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zapatka M., Borozan I., Brewer D.S., Iskar M., Grundhoff A., Alawi M., Desai N., Sültmann H., Moch H., Alawi M., Cooper C.S., Eils R., Ferretti V., Lichter P., Borozan I., Brewer D.S., Cooper C.S., Desai N., Eils R., Ferretti V., Grundhoff A., Iskar M., Kleinheinz K., Lichter P., Nakagawa H., Ojesina A.I., Pedamallu C.S., Schlesner M., Su X., Zapatka M. The landscape of viral associations in human cancers. Nat Genet. 2020;52:320–330. doi: 10.1038/s41588-019-0558-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z., Won H.H., Scott S.N., Brannon R.A., O'Reilly C., Sadowska J., Casanova J., Yannes A., Hechtman J.F., Yao J., Song W., Ross D.S., Oultache A., Dogan S., Borsu L., Hameed M., Nafa K., Arcila M.E., Ladanyi M., Berger M.F. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT) a hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zehir A., Benayed R., Shah R.H., Syed A., Middha S., Kim H.R., et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–713. doi: 10.1038/nm.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y., Ye W., Zhang Y., Xu Y. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 2015;43:7762–7768. doi: 10.1093/nar/gkv784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ondov B.D., Bergman N.H., Phillippy A.M. Interactive metagenomic visualization in a web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shen W., Xiong J. TaxonKit: a cross-platform and efficient NCBI taxonomy toolkit. bioRxiv. 2019. [Preprint] doi: 10.1101/513523. doi: [DOI] [Google Scholar]
- 21.Schache A., Liloglou T., Risk J., Jones T., Ma X.-J., Wang H., Bui S., Luo Y., Sloan P., Shaw R., Robinson M. Validation of a novel diagnostic standard in HPV-positive oropharyngeal squamous cell carcinoma. Br J Cancer. 2013;108:1332. doi: 10.1038/bjc.2013.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang Y.-W., Lozano L., Chen X., Querec T.D., Katabi N., Moreno-Docón A., Wang H., Fix D., Brot L.D., McMillen T.A., Yoon J.-Y., Torroba A., Wang Y., Unger E.R., Park K.J. An isothermal, multiplex amplification assay for detection and genotyping of human papillomaviruses in formalin-fixed, paraffin-embedded tissues. J Mol Diagn. 2020;22:419–428. doi: 10.1016/j.jmoldx.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cho Y., Chang M., Park S., Kim H., Kim W. In situ hybridization of Epstein-Barr virus in tumor cells and tumor-infiltrating lymphocytes of the gastrointestinal tract. Hum Pathol. 2001;32:297–301. doi: 10.1053/hupa.2001.22766. [DOI] [PubMed] [Google Scholar]
- 24.Afzal S., Wilkening S., von Kalle C., Schmidt M., Fronza R. GENE-IS: time-efficient and accurate analysis of viral integration events in large-scale gene therapy data. Mol Ther Nucleic Acids. 2017;6:133–139. doi: 10.1016/j.omtn.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.-C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J., Park W.-Y., Kim N.K., Jang S., Chun S.-M., Sung C.-O., Choi J., Ko Y.-H., Choi Y.-L., Shim H., Won J.-K., Molecular Pathology Study Group of Korean Society of Pathologists Good laboratory standards for clinical next-generation sequencing cancer panel tests. J Pathol Transl Med. 2017;51:191–204. doi: 10.4132/jptm.2017.03.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leal S.M., Gulley M.L. Current and emerging molecular tests for human papillomavirus–related neoplasia in the genomic era. J Mol Diagn. 2017;19:366–377. doi: 10.1016/j.jmoldx.2017.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fernandez S.G., Miranda J. Bendamustine reactivates latent Epstein-Barr virus. Leuk Lymphoma. 2015;57:1208–1210. doi: 10.3109/10428194.2015.1079317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hutt-Fletcher L.M. Epstein–Barr virus replicating in epithelial cells. Proc Natl Acad Sci U S A. 2014;111:16242–16243. doi: 10.1073/pnas.1418974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blair S., Forbes A., Parkins R. CMV colitis in an immunocompetent adult. J R Soc Med. 1992;85:238–239. doi: 10.1177/014107689208500421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Levine M., Laufer I., Kressel H., Friedman H. Herpes esophagitis. Am J Roentgenol. 1981;136:863–866. doi: 10.2214/ajr.136.5.863. [DOI] [PubMed] [Google Scholar]
- 32.Chan A., Bazerbachi F., Hanson B., Alraies M., Duran-Nelson A. Cytomegalovirus hepatitis and pancreatitis in the immunocompetent. Ochsner J. 2014;14:295–299. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z., Aguilar E.G., Luna J.I., Dunai C., Khuat L.T., Le C.T., Mirsoian A., Minnar C.M., Stoffel K.M., Sturgill I.R., Grossenbacher S.K., Withers S.S., Rebhun R.B., Hartigan-O'Connor D.J., Méndez-Lagares G., Tarantal A.F., Isseroff R.R., Griffith T.S., Schalper K.A., Merleev A., Saha A., Maverakis E., Kelly K., Aljumaily R., Ibrahimi S., Mukherjee S., Machiorlatti M., Vesely S.K., Longo D.L., Blazar B.R., Canter R.J., Murphy W.J., Monjazeb A.M. Paradoxical effects of obesity on T cell function during tumor progression and PD-1 checkpoint blockade. Nat Med. 2019;25:141–151. doi: 10.1038/s41591-018-0221-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grulich A.E., van Leeuwen M.T., Falster M.O., Vajdic C.M. Incidence of cancers in people with HIV/AIDS compared with immunosuppressed transplant recipients: a meta-analysis. Lancet. 2007;370:59–67. doi: 10.1016/S0140-6736(07)61050-2. [DOI] [PubMed] [Google Scholar]
- 35.Ihira M., Yoshikawa T., Ohashi M., Enomono Y., Akimoto S., Suga S., Saji H., Nishiyama Y., Asano Y. Variation of human herpesvirus 7 shedding in saliva. J Infect Dis. 2003;188:1352–1354. doi: 10.1086/379040. [DOI] [PubMed] [Google Scholar]
- 36.Gormley R.H., Groft C.M., Miller C.J., Kovarik C.L. Digital squamous cell carcinoma and association with diverse high-risk human papillomavirus types. J Am Acad Dermatol. 2011;64:981–985. doi: 10.1016/j.jaad.2009.10.044. [DOI] [PubMed] [Google Scholar]
- 37.Eliassen E., Lum E., Pritchett J., Ongradi J., Krueger G., Crawford J.R., Phan T.L., Ablashi D., Hudnall S. Human herpesvirus 6 and malignancy: a review. Front Oncol. 2018;8:512. doi: 10.3389/fonc.2018.00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pellett P.E., Ablashi D.V., Ambros P.F., Agut H., Caserta M.T., Descamps V., Flamand L., Gautheret-Dejean A., Hall C.B., Kamble R.T., Kuehl U., Lassner D., Lautenschlager I., Loomis K.S., Luppi M., Lusso P., Medveczky P.G., Montoya J.G., Mori Y., Ogata M., Pritchett J.C., Rogez S., Seto E., Ward K.N., Yoshikawa T., Razonable R.R. Chromosomally integrated human herpesvirus 6: questions and answers. Rev Med Virol. 2012;22:144–155. doi: 10.1002/rmv.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lazo P., DiPaolo J., Popescu N. Amplification of the integrated viral transforming genes of human papillomavirus 18 and its 5’-flanking cellular sequence located near the myc protooncogene in HeLa cells. Cancer Res. 1989;49:4305–4310. [PubMed] [Google Scholar]
- 40.Furuta M., Tanaka H., Shiraishi Y., Unida T., Imamura M., Fujimoto A., Fujita M., Sasaki-Oku A., Maejima K., Nakano K., Kawakami Y., Arihiro K., Aikata H., Ueno M., Hayami S., Ariizumi S.-I., Yamamoto M., Gotoh K., Ohdan H., Yamaue H., Miyano S., Chayama K., Nakagawa H. Characterization of HBV integration patterns and timing in liver cancer and HBV-infected livers. Oncotarget. 2018;9:25075–25088. doi: 10.18632/oncotarget.25308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yeh Y.-C., Kao H.-L., Lee K.-L., Wu M.-H., Ho H.-L., Chou T.-Y. Epstein-Barr virus–associated pulmonary carcinoma. Am J Surg Pathol. 2019;43:211–219. doi: 10.1097/PAS.0000000000001173. [DOI] [PubMed] [Google Scholar]
- 42.Vandeven N., Nghiem P. Rationale for immune-based therapies in Merkel polyomavirus-positive and -negative Merkel cell carcinomas. Immunotherapy. 2016;8:907–921. doi: 10.2217/imt-2016-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lassen P., Eriksen J.G., Hamilton-Dutoit S., Tramm T., Alsner J., Overgaard J. Effect of HPV-associated p16INK4A expression on response to radiotherapy and survival in squamous cell carcinoma of the head and neck. J Clin Oncol. 2009;27:1992–1998. doi: 10.1200/JCO.2008.20.2853. [DOI] [PubMed] [Google Scholar]
- 44.Bzhalava Z., Tampuu A., Bała P., Vicente R., Dillner J. Machine learning for detection of viral sequences in human metagenomic datasets. BMC Bioinformatics. 2018;19:336. doi: 10.1186/s12859-018-2340-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soueidan H., Nikolski M. Machine learning for metagenomics: methods and tools. arXiv. 2015 doi: 10.48550/arXiv.1510.06621. doi: [DOI] [Google Scholar]
- 46.Suh N., Liapis H., Misdraji J., Brunt E.M., Wang H.L. Epstein-Barr virus hepatitis: diagnostic value of in situ hybridization, polymerase chain reaction, and immunohistochemistry on liver biopsy from immunocompetent patients. Am J Surg Pathol. 2007;31:1403–1409. doi: 10.1097/PAS.0b013e31802ffdd5. [DOI] [PubMed] [Google Scholar]
- 47.Abreu A.L., Souza R.P., Gimenes F., Consolaro M.E. A review of methods for detect human papilloma virus infection. Virol J. 2012;9:262. doi: 10.1186/1743-422X-9-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duncavage E.J., Le B.-M., Wang D., Pfeifer J.D. Merkel cell polyomavirus. Am J Surg Pathol. 2009;33:1771–1777. doi: 10.1097/PAS.0b013e3181ba7b73. [DOI] [PubMed] [Google Scholar]
- 49.Louis C.U., Straathof K., Bollard C.M., Ennamuri S., Gerken C., Lopez T.T., Huls H.M., Sheehan A., Wu M.-F., Liu H., Gee A., Brenner M.K., Rooney C.M., Heslop H.E., Gottschalk S. Adoptive transfer of EBV-specific T cells results in sustained clinical responses in patients with locoregional nasopharyngeal carcinoma. J Immunother. 2010;33:983–990. doi: 10.1097/CJI.0b013e3181f3cbf4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Virus tumor associations in validation cohort. A: Forest plot demonstrating the statistically significant (P < 0.005) virus-tumor type relationships that are enriched in the validation cohort of 7813 consecutive samples using the updated version of the assay with virus-specific probes. Error bars indicate the 95% confidence interval. Associations are arranged by virus. B: Photomicrogaphs of a case of digital papillary adenocarcinoma (DPA). Scale bar = 100 micrometers. The top panel indicates H&E and the bottom panel indicates an HPV ISH using RNAScope probes specific to the HPV42 RNA, designed internally to assist with clinical identification of DPA based on the association established by this study.
Microbiome Extraction Pipeline
Stastical Analysis