Fig. 1 |. Overview of the trigenic SGA (τ-SGA) screening and quantification methodology.
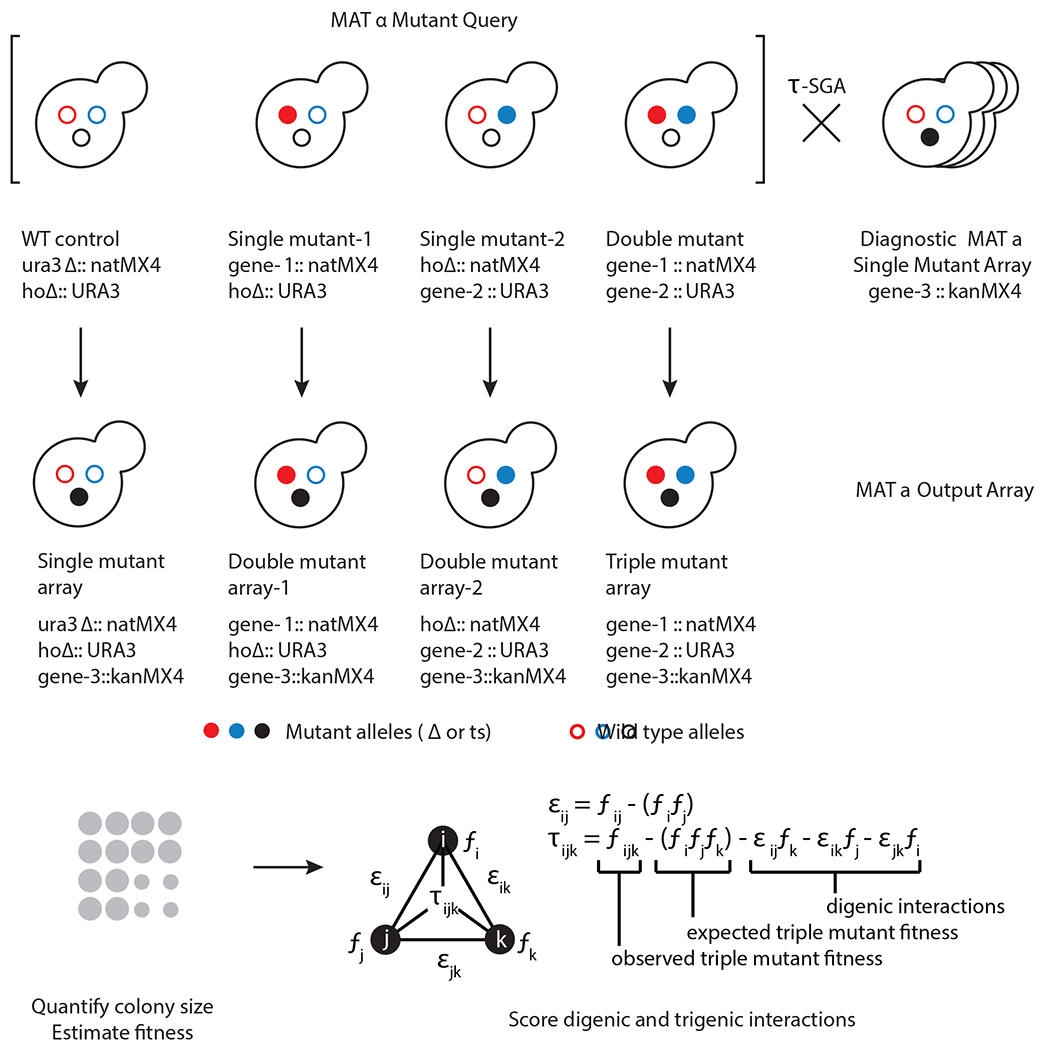
Top, a double mutant query strain with its two corresponding single mutant query strains and a wild-type reference strain are crossed in parallel using τ-SGA automated replica pinning and selection steps to the diagnostic array, which is representative of the genome-wide array collection to generate respective triple, double and single mutants carrying the desired genetic markers. Bottom, τ-SGA scoring pipeline is used to quantify trigenic interactions using colony size as a proxy for fitness accounting for any digenic interactions.
