Fig. 2 |. Classes of trigenic interactions.
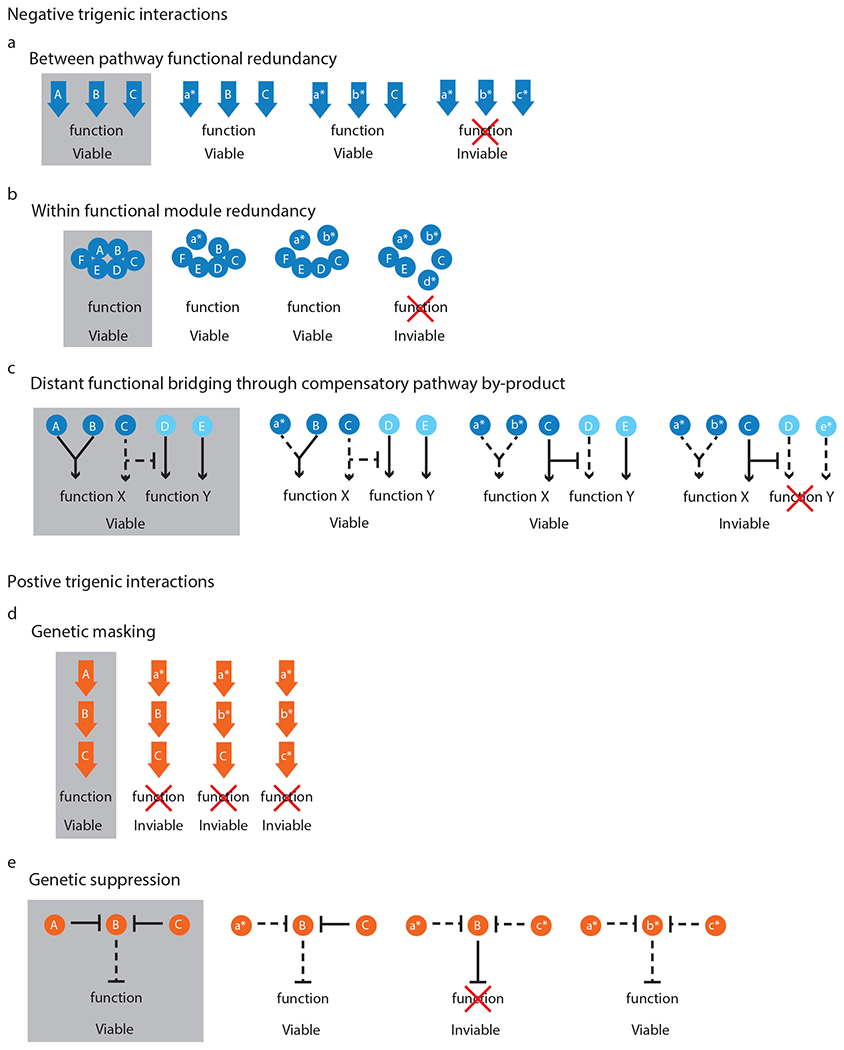
a, Between-pathway functional redundancy, when any of the three genes (A-C) are sufficient for pathway function e.g. ODC1, ODC2 and CTP1 or CLN1, CLN2 and CLN3; b, within functional module redundancy, when a protein complex contains up to three non-essential subunits in which the disruption of any combination of two components is tolerated, but a third perturbation results in sickness (A, B, D) e.g. OCA4, OCA5 and OCA6 or SET1, SDC1 and SWD1; c, distant functional bridging through compensatory pathway by-product, when the activity of two genes (A, B e.g. MDY2, MTC1) impinges on a common function (e.g. trafficking of aromatic amino acid transporters) and their perturbation activates a compensatory pathway (C e.g. de novo production of aromatic amino acids) that generates a by-product (e.g. NAD+) that impairs another function (e.g. telomere capping), which is mediated by another gene (D), which itself is synthetic lethal with another parallel pathway (E e.g. DNA damage repair pathway, MRE11); d, genetic masking of a multi-member pathway, when the perturbation of each individual gene is detrimental to cell fitness, but a second or a third perturbation does not increase the disruptive effect (A-C e.g. RAD52, RAD51 and RAD54); e, genetic suppression is observed when a double mutant is sick (A, C e.g. CAC1 and HIRA complex member), but a perturbation in a third gene (B e.g. ASF1) suppresses the negative effect which could be caused by a third gene (B) whose function becomes toxic in the absence of the other two genes (A, C). An uppercase letter denotes a wild-type allele, a lowercase letter with an asterisk denotes a mutant allele, a solid line denotes an active pathway and dashed line denotes a pathway, which is inactive or reduced in its activity.
