Fig. 4 |. Trigenic Synthetic Genetic Array (τ-SGA) experimental pipeline.
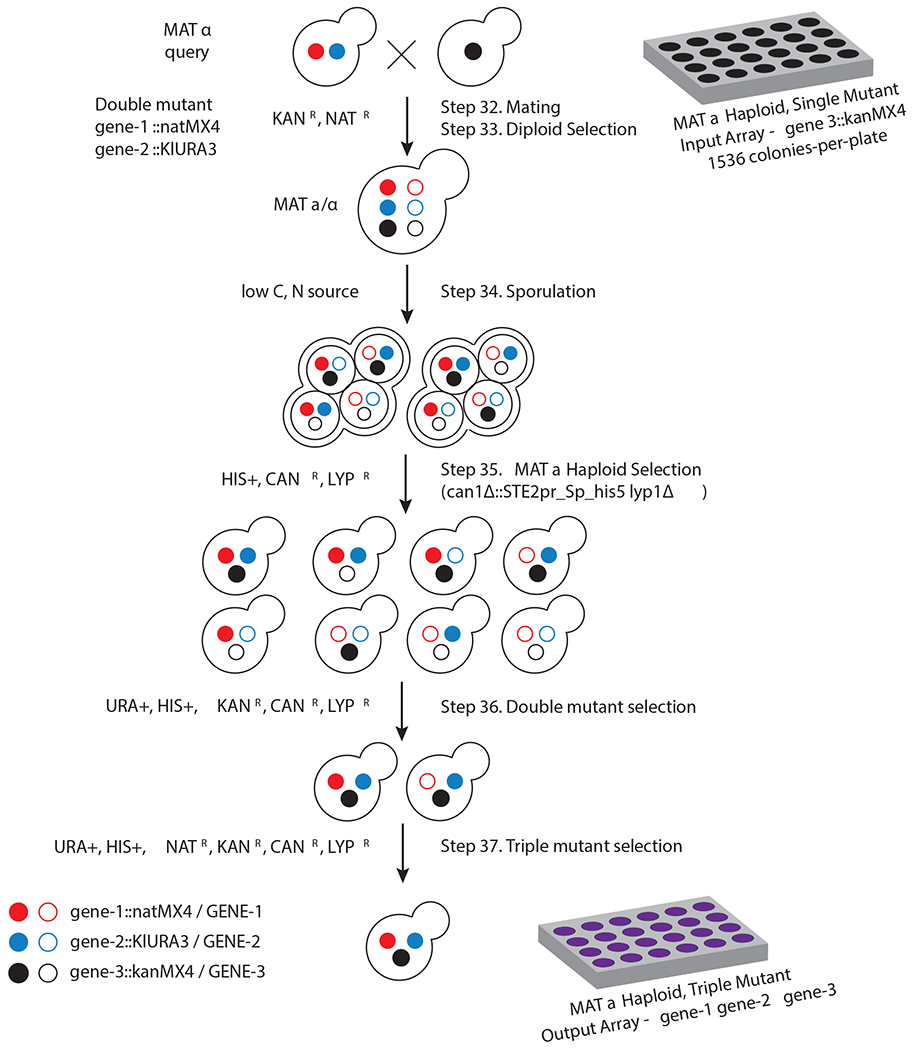
A MATα query double mutant strain harboring loss of function mutations in two different genes of interest linked to dominant selectable markers (natMX4 and KIURA3), which confer resistance to the antibiotic nourseothricin and the ability to grow in media lacking uracil (filled red and blue circles, respectively). The arginine and lysine permeases, CAN1 and LYP1, which confer sensitivity to canavanine and thialysine, are also deleted in the query strain and are used to select against diploids following the sporulation step. Typically, in a τ-SGA screen the query strain is crossed to an ordered diagnostic array of MATa non-essential gene deletion strains and ts alleles of essential genes (‘array’ mutants) linked to a dominant selectable marker, kanMX4, which confers geneticin resistance (filled black circle). The resulting heterozygous diploids are replica-pinned to a medium low in carbon and nitrogen sources to induce meiosis. The resulting sporulated mix is then replica-pinned to a synthetic medium depleted for histidine, but containing canavanine and thialysine to allow for the selective germination of MATa haploid meiotic progeny. This selective germination is possible because the SGA reporter, STE2pr_Sp_his5, in which the STE2 MATa-specific promoter (STE2pr) controls the expression of the Schizosaccharomyces pombe his5 gene, replaces the CAN1 gene. The MATa haploid progeny is then transferred to the selective medium lacking uracil and containing geneticin to select for mutants harboring one of the query strain mutations and an array mutation. In the final step, the selected haploids are transferred to the medium lacking uracil and containing geneticin and nourseothricin to select for all three markers.
