Figure 5.
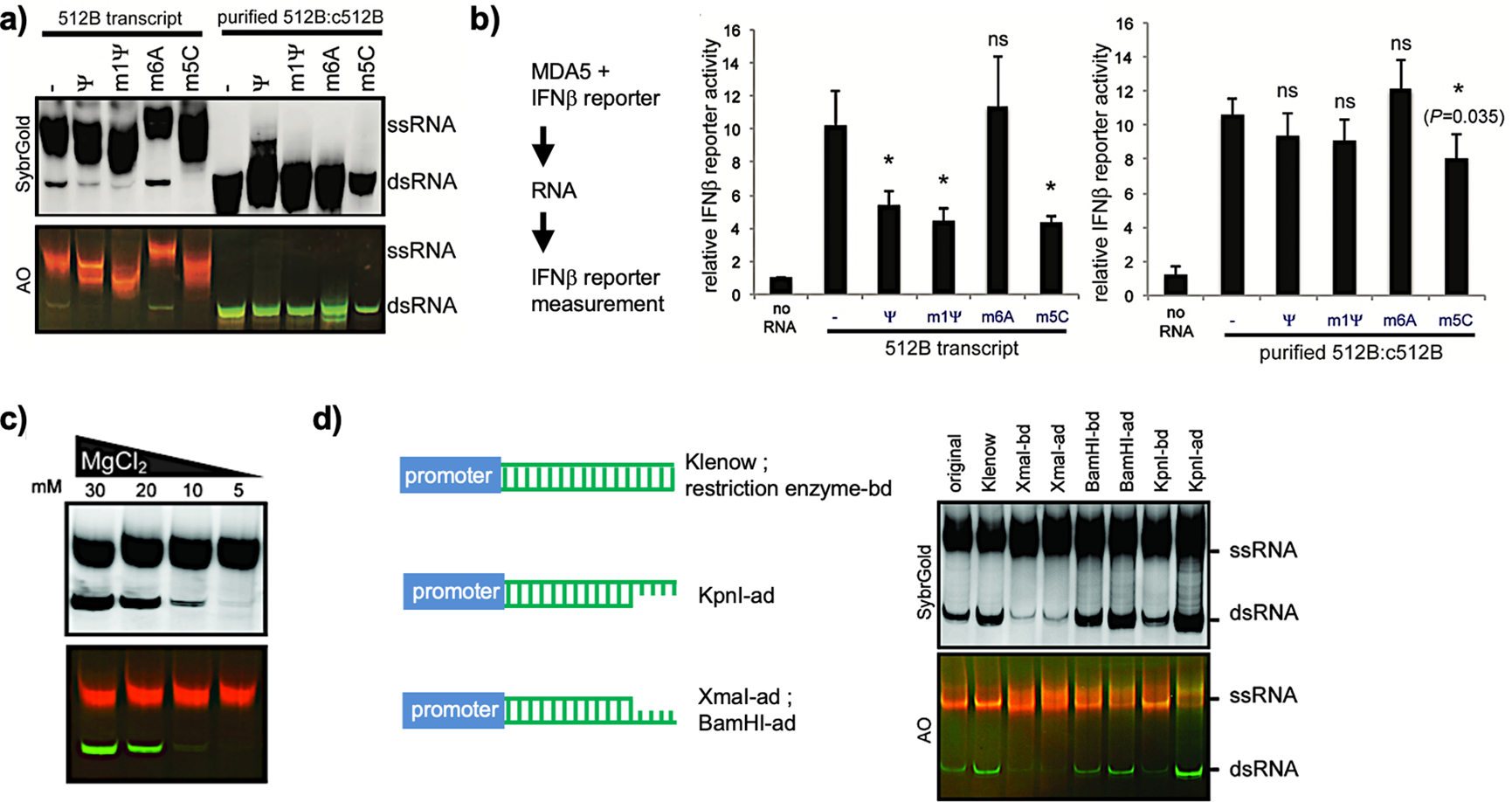
(a) Native PAGE analysis of transcripts using unmodified or modified nucleotides for IVT. The purified dsRNA 512B:c512B was initiated by incubation of IVT products with RNase I to digest ssRNAs. (b) A luciferase assay was done in HEK293T cells by cotransfection of MDA5-expressing plasmids and luciferase reporters driven by IFNβ promoter, followed by unpurified (512B transcript) or purified (purified 512B:c512B) RNA transfection, respectively. (c) Native PAGE analysis of transcribed RNAs at 512 nucleotides (nt) using different Mg2+ concentrations for IVT. dsRNA byproducts were validated by AO staining. (d) Native PAGE analysis of transcribed RNAs with different 3′-end sequences and structures. Templates before (original) and after the Klenow reaction (Klenow), derivatives with indicated restriction sites at the 3′-end before digestion (-bd) and after digestion (-ad) are shown on the left and were used for IVT on the right. (a–d) Reprinted with permission from ref 4. Copyright 2018 Oxford University Press.
