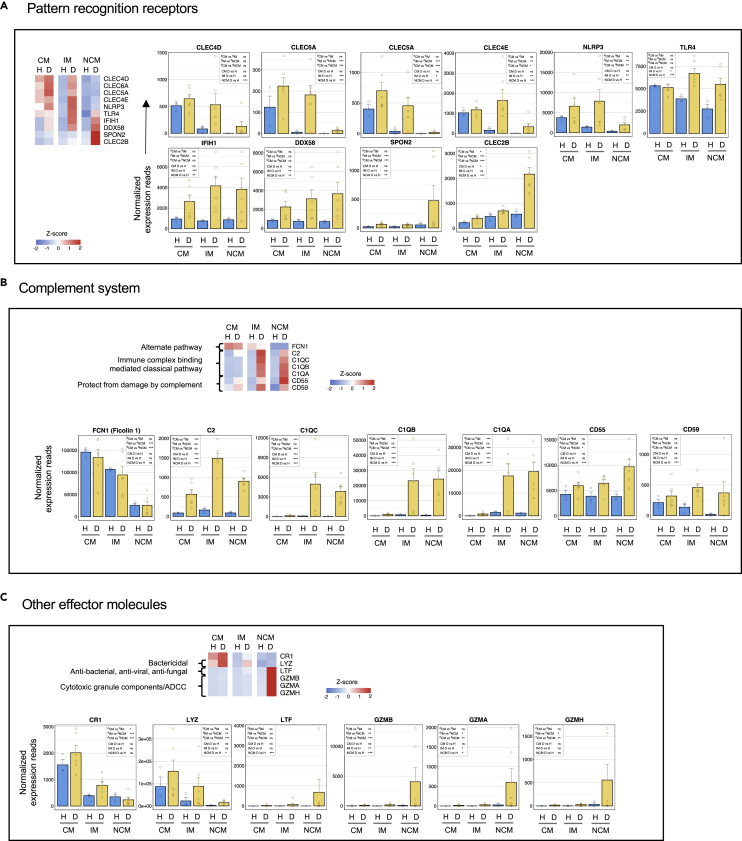
Figure 6.
The CM and IM subsets from dengue patients are biased toward an expression of key genes involved in microbial carbohydrate pattern recognition and inflammation, the IM and NCM are biased toward intracellular double-stranded RNA recognition – whereas the NCM are biased toward pattern recognition in the extracellular matrix and monocyte-NK cross talk
(A) Select genes related to pattern recognition.
(B) Select genes related to complement.
(C) Select genes related to other effector functions. For heatmaps, the gradient of high to low gene expression based on Z-score calculated from the average of normalized counts for healthy subjects and dengue patients in each monocyte subset is shown from red to blue. Panels of bar graphs showing normalized expression read in CM, IM, and NCM subsets from healthy children and dengue patients are grouped as prerecording to their bias toward CM, IM, or NCM. Each dot represents the normalized counts for the individual subject (healthy, n = 3; dengue, n = 6). Bars represent average reads in a given subset for a given condition. Blue: healthy children; Yellow: dengue febrile children. Error bars represent the SEM. For each bar graph, significant and non-significant differences within subsets from dengue patients (dCM, dIM and dNCM) and for dengue versus healthy comparison (D vs H) in each subset (CM, IM and NCM) is highlighted in a box, where ∗ denotes adjusted P-value <0.05; ∗∗ denotes adjusted P-value <0.01; ∗∗∗ denotes adjusted P-value <0.001; and “ns” denotes non-significant (also mentioned in Table S3G). The adjusted p values were obtained from DeSeq2 analysis.