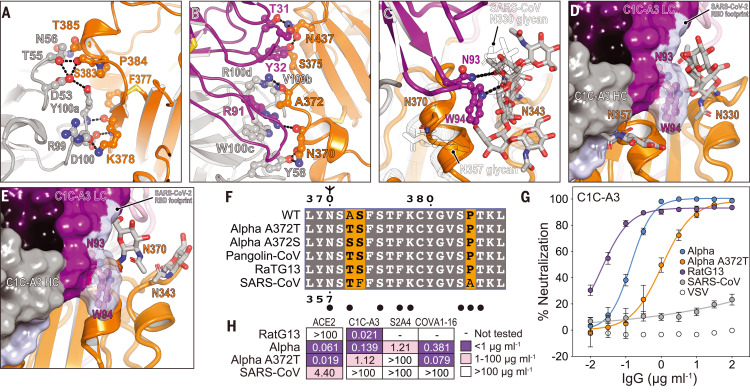
Fig. 5. Structural basis for immune evasion of a RBD core–targeting antibody.
(A and B) C1C-A3 antibody contacts with the SARS-CoV-2 RBD core. (C) C1C-A3 contacts with the N343RBD glycan with structural superposition of the SARS-CoV RBD (PDB ID 6NB6) (78). N-linked glycans found at N330RBD and N357RBD in SARS-CoV and the analogous N343RBD and N370RBD positions in SARS-CoV-2 are highlighted. (D) Superposition of the C1C-A3 Fab–SARS-CoV-2 RBD structure with the SARS-CoV RBD (PDB ID 6NB6) (78) showing that a glycan attached at SARS-CoV N357RBD may interfere with antibody binding. The SARS-CoV-2 RBD is not shown for clarity. (E) Superposition of the C1C-A3–SARS-CoV-2 RBD with the RaTG13 virus RBD (PDB ID 7CN4) (79) showing that a glycan attached at RaTG13 virus N370RBD would be more readily accommodated because the helix that contains it would be rotated away from the Fab. The SARS-CoV-2 RBD is omitted for clarity. (F) Sequence alignment of the RBD core region contacted by C1C-A3. SARS-CoV-2 numbering is shown at the top of the alignment, and SARS-CoV numbering is shown at the bottom. Circles indicate antibody contacts. (G) C1C-A3 neutralization curves for the indicated lentivirus pseudotypes. Data are plotted as the mean ± standard deviation of the mean. The experiment was performed twice in triplicate (n = 6). For some data points, error bars are smaller than symbols. (H) Tabulated neutralization IC50 values for the indicated pseudotypes.