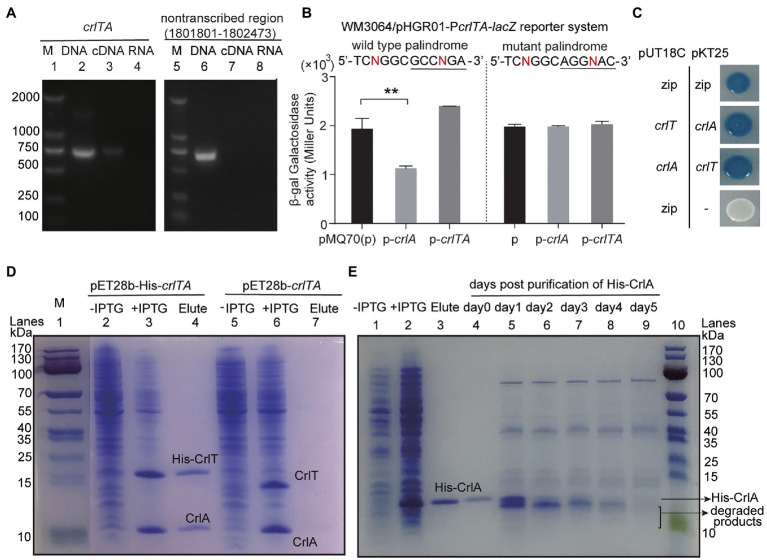
Figure 3.
Characteristics of the type II TA pair CrlTA. (A) Total gDNA and RNA were extracted from P. aeruginosa WK172, and total RNA was used to synthesize cDNA. PCR was carried out by primer pairs crlT-F/crlA-R and non-transcribed region-F/R using gDNA (lanes 2, 5), cDNA (lanes 3, 6), and RNA (lanes 4, 7). The DNA marker (M) is in lane 1. (B) Mid-log phase WM3064 cells carrying reporter plasmids pHGR01-PcrlTA-lacZ or pHGR01-MPcrlTA-lacZ (M indicates mutant) were transfected with pMQ70-based plasmids. The double plasmid systems were used to test β-galactosidase activity with induction of 10 mM arabinose for 3 h. Student’s t-test was used for statistical analysis and error bars indicate the standard error of the mean (n = 3, p < 0.01 is shown in **). (C) CrlT and CrlA were fused to the T18 catalytic domain in pUT18C and the T25 catalytic domain in pKT25, respectively. In contrast, CrlT and CrlA were also fused to T25 and T18 in the corresponding pKT25 and pUT18C. Cells harboring pKT25-zip and pUT18C-zip plasmids were used as positive controls, and cells harboring pKT25 (without an insert) and pUT18C-zip plasmids were used as negative controls. (D) NHis-tagged CrlT and untagged CrlA were produced from pET28b-His-crlTA in E. coli BL21. CrlA (11.92 kDa) was copurified with NHis-CrlT (14.73 kDa, lane 4). Cells harboring pET28b-crlTA were used as a negative control (lane 7). The protein marker (M) was loaded in lane 1. (E) NHis-tagged CrlA was purified and stored at 4°C, and the stability of the purified protein was determined each day after purification.