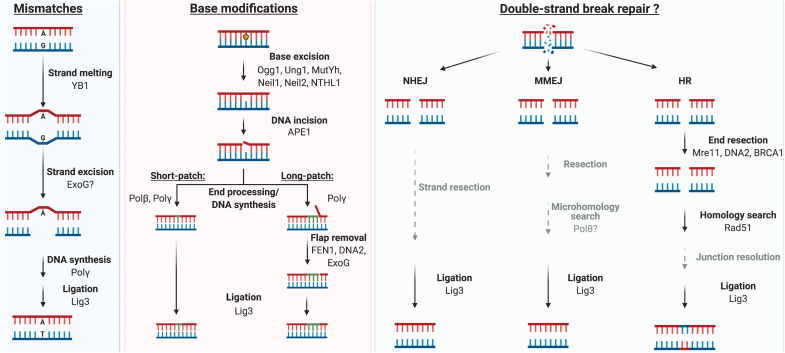
FIG. 5.
Schematic representation of putative mtDNA repair pathways. For the MMR (left panel), YB-1 is the only mitochondrial MMR-specific protein identified. The ExoG nuclease is found in mitochondria, but its role in MMR still lacks some evidence. BER (middle panel) responds to small base modifications, and proteins for all steps of this pathway are found in mitochondria. A range of DNA glycosylases with distinct substrate specificities can excise the damaged base; some glycosylases such as OGG1 can also incise the DNA backbone to allow excision of the AP-site. DNA synthesis occurs after end processing (details not shown). Long-patch BER involves nucleases such as FEN1, DNA2, or ExoG to remove the flap generated during strand displacement synthesis. DSB repair (right panel) has not been conclusively reported in mitochondria, but in the nucleus it occurs via HR, NHEJ, or its sub-pathway, MMEJ. Some factors of these DSB repair pathways, such as Polθ that may act in MMEJ or the HR factor Rad51, and end resection proteins Mre11, DNA2, and BRCA1 are found in mitochondria, although their roles in this organelle remain elusive. Intermediate steps that lack evidence are in gray. In all repair pathways, LIG3 is believed to catalyze the ligation step. HR, homologous recombination; LIG3, ligase 3; MMR, mismatch repair; NHEJ, non-homologous end joining; OGG1, 8-oxoguanine glycosylase. Color images are available online.