Figure 3.
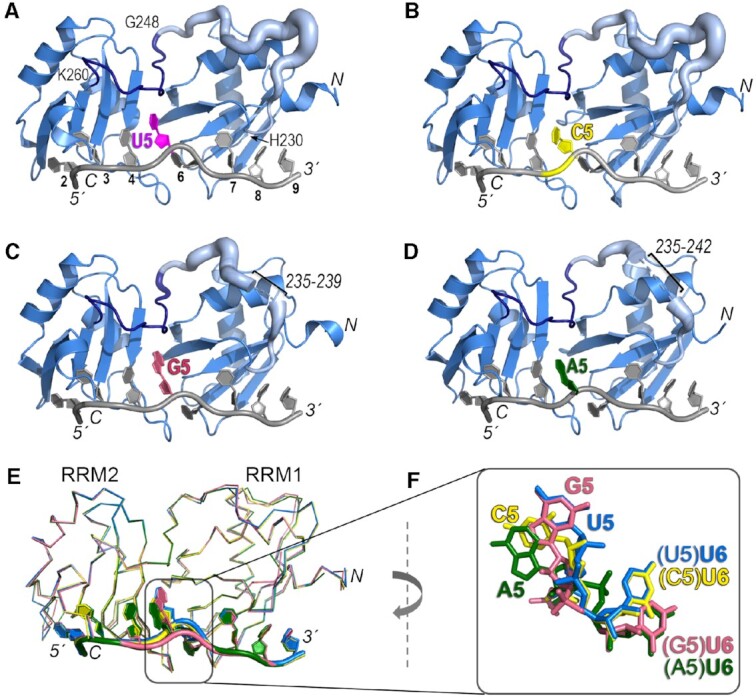
Crystal structures of U2AF212L recognizing AdML Py tract variants that differ in the identities of the central nucleotide. The amino (N)/carboxy (C)-termini of the polypeptide and 5′/3′ termini of the oligonucleotide are labeled in italics. The nucleotide positions are numbered on panel A. (A–D) Overall ribbon diagrams of the protein (blue) bound to oligonucleotides (grey) substituted with (A) uridine (U5, magenta, PDB ID 6XLW), (B) cytidine (C5, yellow, PDB ID 7S3A, this study), (C) guanosine (G5, salmon, PDB ID 7S3B, this study), or (D) adenosine (A5, green, PDB ID 7S3C, this study). On panels A–D, the temperature factors (mobility) of the inter-RRM linker (residues 230–260) are represented using cartoon putty in PyMOL, which scales the size of the coil proportionately to the temperature factors of the residues. Residues 230–247 are pale blue and residues 248–260 are dark blue (boundary residues labeled on panel A). Ranges of linker residues that unresolved in the G5 and A5 structures are labeled. (E) Superposition by matching Cα atoms in the four structures shows conformational changes at the fifth and sixth nucleotide positions (numbered according to the nine nucleotide-binding sites of PDB ID 5EV4). (F) Closer view of the fifth and sixth nucleotides shown in (E) following a 90° clockwise rotation about the y-axis relative to (E).
