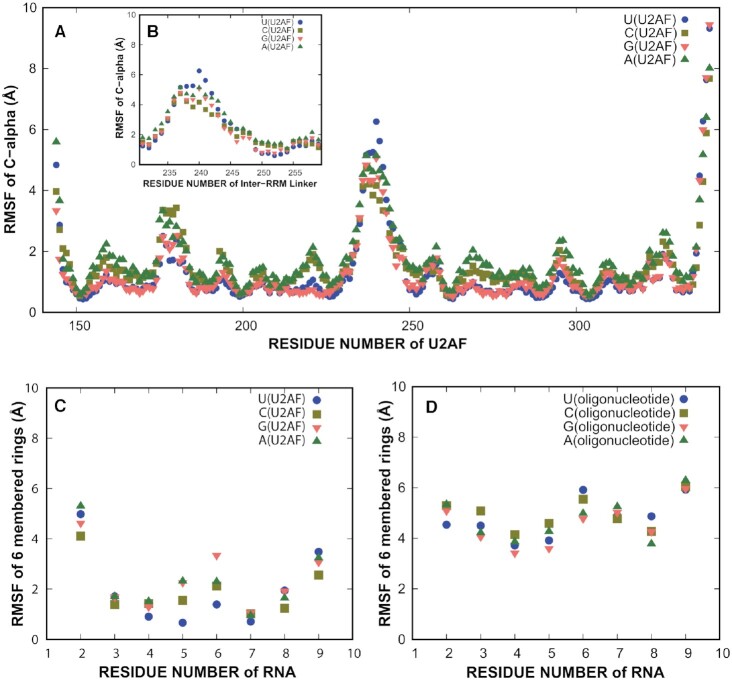
Figure 5.
Molecular dynamics studies of conformational flexibility. (A) Root mean squared fluctuations (RMSF) of U2AF2 Cα by residue numbers. The RRM1 and RRM2 regions are relatively static. (B) Inset showing RMSF of the inter-RRM linker Cαs (residues 230–260). (C) RMSF values of six-membered rings of the RNA in the U2AF2-RNA complex simulation (2 μs). (D) RMSF values of six-membered rings of the RNA in the oligonucleotide simulations (1 μs). The oligonucleotide simulations have higher fluctuations than the U2AF2–RNA simulations. Supplementary Figures S4 and S5 show RMSDs to demonstrate stability of the protein during simulation and convergence of the simulation.