Figure 2:
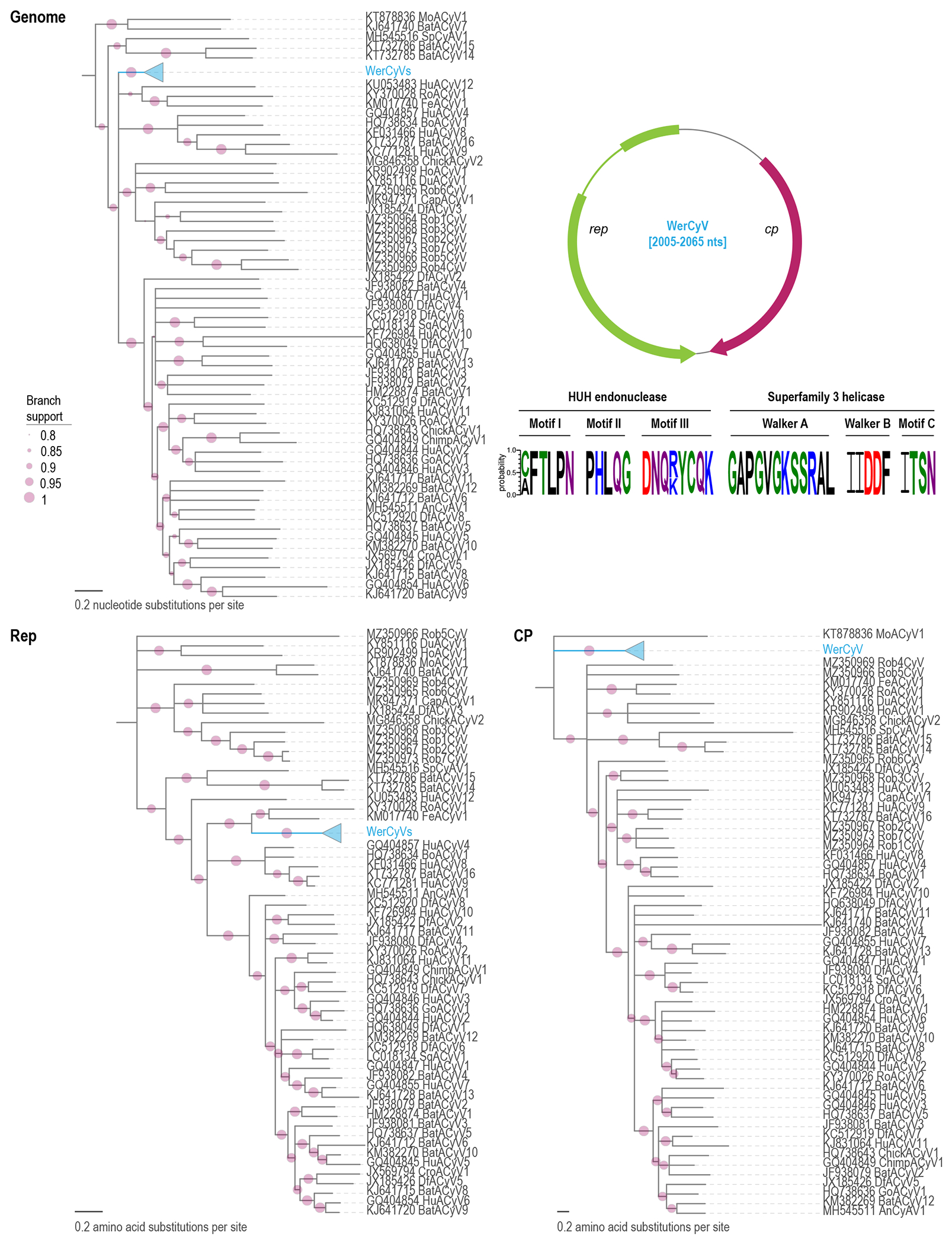
Maximum-likelihood phylogenetic tree of genome sequences of cycloviruses, and the Rep and CP amino acid sequences. The 45 WerCyV sequences are shown as a collapsed clade. An expansion of this clade is shown in Figure 3. The phylogenetic trees have been rooted with representative sequences of circoviruses and branches with <0.8 aLRT branch support have been collapsed with TreeGraph2 (Stover and Muller, 2010). On the top right the genome organization of WerCyV is illustrated. The conserved motifs in the Rep of WerCyV are shown with Weblogo (Crooks et al., 2004) and the residues are colored based on their chemical properties (acidic, red; basic, blue; hydrophobic, black; neutral, purple; polar, green).
