Figure 5.
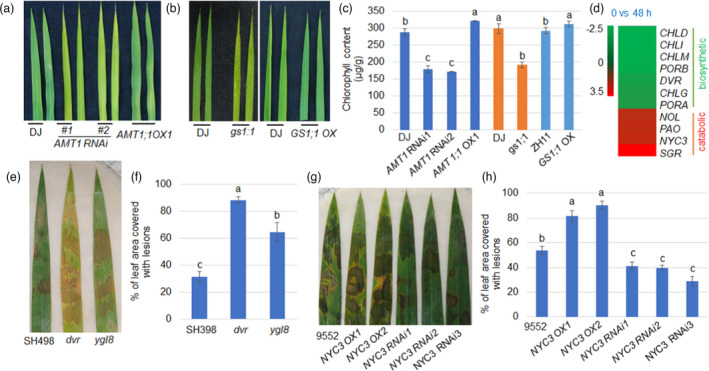
Chlorophyll accumulation promotes rice defence to ShB. (a) The leaves of AMT1 RNAi (#1 and #2), AMT1;1 OX and wild‐type were photographed. (b) Leaves from gs1;1, GS1;1 OX together with their corresponding wild‐type were photographed. (c) The chlorophyll contents of AMT1 RNAi, AMT1;1 OX and GS1;1 mutants as well as their corresponding wild‐type were determined. (d) R. solani AG1‐IA dependent (after 48 hours of inoculation) expression levels of chlorophyll biosynthesis and catabolic genes were shown in a heat map. (e) The response of chlorophyll synthesis‐related gene DVR and YGL8 mutants and wild‐type Shuhui498 (SH498) to R. solani. (f) The lesion scales were analysed for the R. solani AG1‐IA‐infected leaves shown in (e) by determination of the lesion area on the leaf surface. (n > 10). (g) The response of chlorophyll catabolic gene NYC3 RNAi and overexpression (OX) plants to R. solani. (h) The corresponding lesion scales were analysed by determination of the lesion area on the leaf surface. Data represent means ± standard error (SE) (n > 10). Significant differences at the P < 0.05 level are indicated by different letters.
