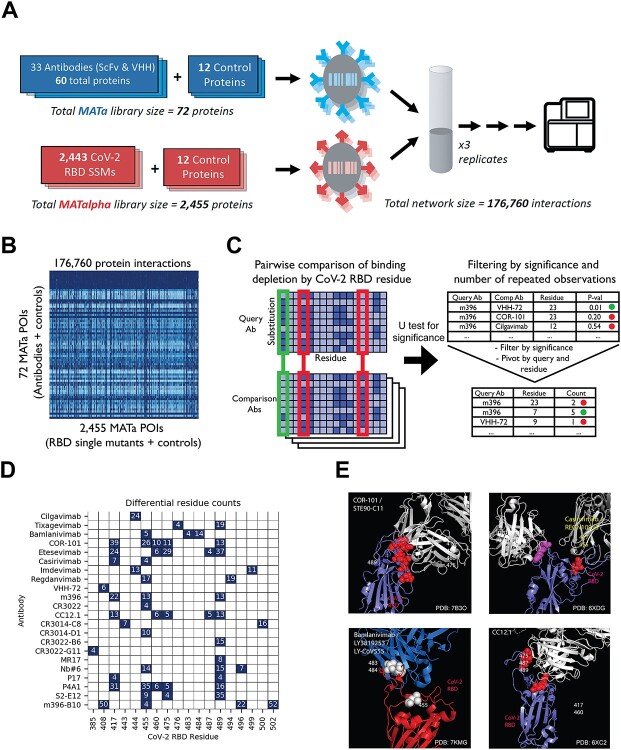
Figure 2.
(A) Schematic of epitope mapping AlphaSeq experiment, comparing antibodies against a CoV-2 RBD SSM library. (B) Heatmap of AlphaSeq binding data showing all interactions measured in the assay in log10 KD (nM). RBD–CoV-2 intolerant substitutions results in loss binding for all antibodies in the set and appear as vertical dark blue streaks. (C) Method summary for determination of epitope residues; each antibody was compared against all others, and at each RBD site a Mann–Whitney U test was performed to determine if binding was more impacted by a RBD mutations at that site in one antibody; results were filtered by U test significance and for significance in four or more pairwise comparisons. (D) Summary of epitope determination results. A putative epitope residue is represented as a navy color in the figure. An RBD residue is determined to be a putative epitope residue if binding was found to be significantly impacted by substitutions in that residue relative to at least four other antibodies in the dataset. This comparison is necessary to differentiate between substitutions that ablate binding to all antibodies, likely due to misfolding or low expression, versus substitutions that decrease binding to only a fraction of antibodies and therefore are more likely to be at the binding interface. The differential residue counts indicate the number of comparison antibodies for which a relative difference was found for a given antibody and RBD residue (summing the results from light-heavy and heavy-light orientations when both were tested). (E) Representative antibodies (in blue) with known structure binding to RBD (in red), with residues called as epitope locations marked in white or yellow.