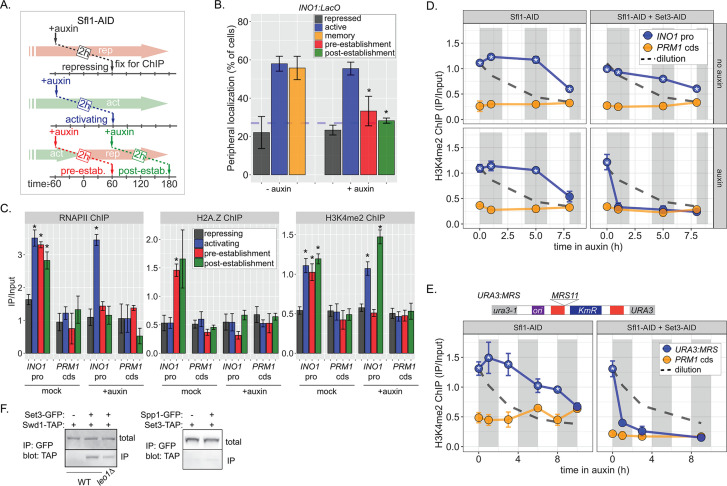
Figure 6. Distinct molecular requirements for establishment and inheritance of H3K4me2 during INO1 memory.
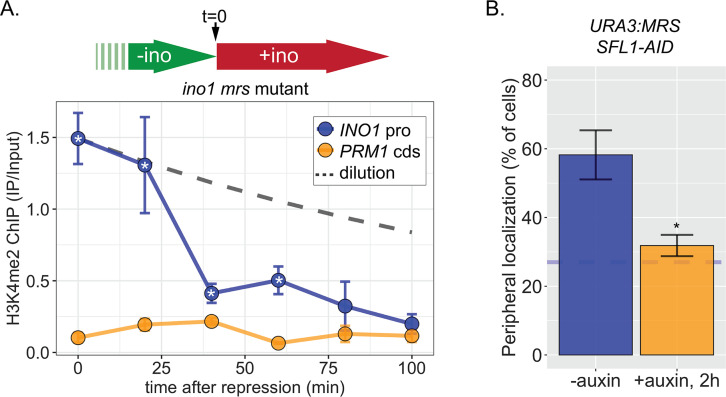
(A) Experimental set-up to test the role of Sfl1 in establishment and inheritance of INO1 memory, using auxin-inducible degradation of Sfl1 before or after establishing memory. The top arrow indicates when auxin was added; the bottom arrow indicates when cells were fixed for chromatin immunoprecipitation (ChIP). Using this approach, peripheral localization of INO1 (B) or ChIP against RNAPII (C, left), H2A.Z (C, middle), or H3K4me2 (C, right) was measured under the indicated conditions ±0.5 mM auxin. Peripheral localization is the average of three biological replicates ± SEM; each biological replicate ≥30 cells. Blue hatched line: expected peripheral localization for a randomly localized gene. *p<0.05 from one-tailed t-test comparing auxin treated to untreated, alternative = less. (D) ChIP against H3K4me2 in Sfl1-AID (left) and Sfl1-AID+Set3 AID (right) strains either without auxin (top) or after addition of auxin (lower). Auxin was added after 1 hr of repression. For panels D and E, vertical gray and white bars represent estimated generation times and dashed line represents the expectation from perfect retention of H3K4me2, followed by dilution through DNA replication (i.e. t1/2 = the doubling time). (E) Schematic of insertion of 11 bp MRS at the URA3 locus in the URA3:MRS strain (top) and ChIP against H3K4me2 in URA3:MRS Sfl1-AID (left) and URA3:MRS Sfl1-AID+Set3 AID (right) strains after addition of auxin (lower). Panels C, D, and E: recovery of INO1 promoter or PRM1 coding sequence or URA3:MRS region was quantified by quantitative PCR relative to input and are the averages of three biological replicates ± SEM. *p<0.05 from one-tailed t-test comparing to recovery of each DNA in the repressed condition (C) or to PRM1 cds (D & E), alternative = greater. (F) Co-immunoprecipitation of Set3-GFP and Swd1-TAP (left), Spp1-GFP and Set3-TAP (right) from the indicated strains. The Green fluorescent protein (GFP)-tagged proteins were immunoprecipitated with anti-GFP nanobodies; recovery of Swd1-TAP and Set3-TAP were monitored by immunoblotting with anti-TAP antibody.