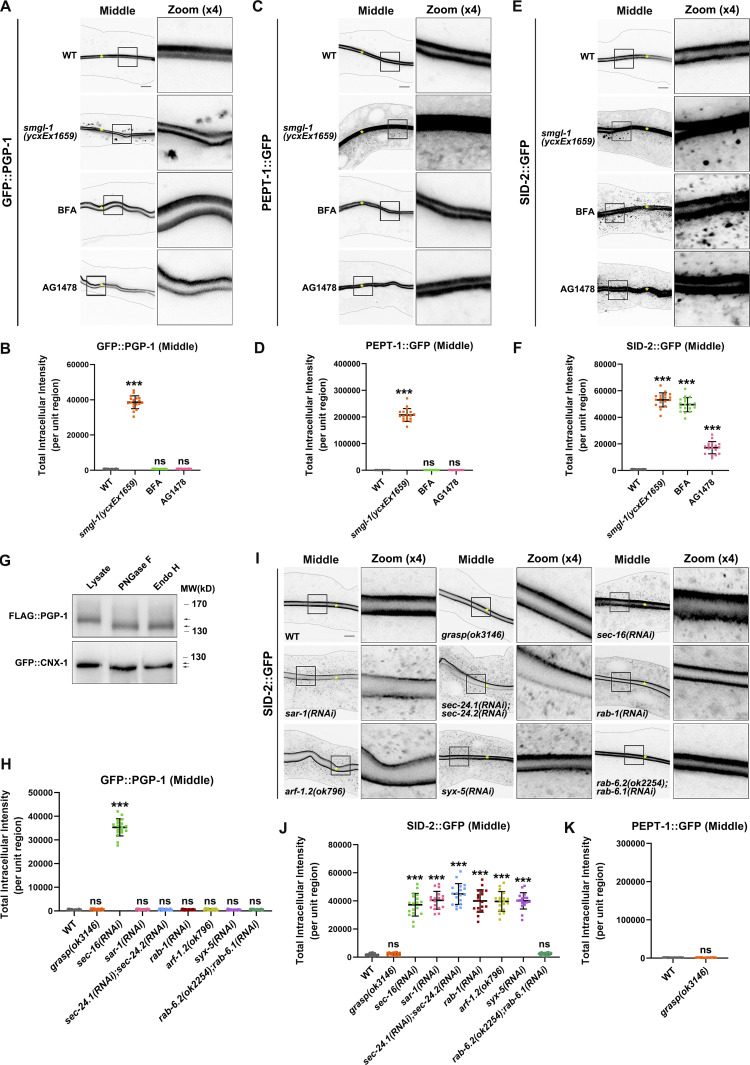
Figure 2.
SMGL-1 regulates the Golgi-bypassing unconventional secretion of apical integral proteins. (A–D) Confocal images of intestinal cells expressing GFP-tagged PGP-1 (A and B) and PEPT-1 (C and D). Compared with wild-type animals, smgl-1(ycxEx1659) mutants displayed intracellular accumulation of GFP::PGP-1 and PEPT-1::GFP. In contrast, BFA and AG1478 treatment did not affect the apical localization of GFP::PGP-1 and PEPT-1::GFP. (E and F) Confocal images of intestinal cells expressing GFP-tagged SID-2. Compared with wild-type animals, smgl-1(ycxEx1659) mutants displayed intracellular accumulation of SID-2::GFP. BFA and AG1478 treatment also caused intracellular accumulation of SID-2::GFP. (G) FLAG::PGP-1 and GFP::CNX-1 transgenic animals were analyzed by immunoblotting using anti-FLAG and anti-GFP antibodies after Endo H or PNGase F treatment. The molecular weight of FLAG::PGP-1 and GFP::CNX-1 is decreased (lower arrow) after Endo H treatment, compared with control animals (upper arrow). CNX-1, a C. elegans homolog of Calnexin, is an ER-resident glycoprotein and Endo H sensitive. Thus, CNX-1 was deployed as a positive control of Endo H treatment. (H) The apical localization of GFP::PGP-1 did not change in animals deficient in GRASP, SAR-1/Sar1, SEC-24.1/Sec24C/Sec24D and SEC-24.2/Sec24A/Sec24B, RAB-1/Rab1, ARF-1.2/Arf1, SYX-5/Stx5, RAB-6.1 and RAB-6.2. In contrast, SEC-16/Sec16 knockdown caused intracellular accumulation of GFP::PGP-1. (I and J) Confocal images of intestinal cells expressing GFP-tagged SID-2. The apical localization of SID-2::GFP did not change in animals deficient in GRASP or RAB-6.1 and RAB-6.2. However, SID-2::GFP displayed abnormal localization in animals deficient in SEC-16/Sec16, SAR-1/Sar1, SEC-24.1/Sec24C/Sec24D and SEC-24.2/Sec24A/Sec24B, RAB-1/Rab1, ARF-1.2/Arf1, and SYX-5/Stx5. (K) GRASP knockdown did not cause intracellular accumulation of PEPT-1::GFP. The signals from the apical membrane were avoided by manual ROI selection. Data are shown as mean ± SD (n = 18 each, six animals of each genotype sampled in three different unit regions of each intestine defined by a 100 × 100 [pixel2] box positioned at random). Statistical significance was determined using a two-tailed, unpaired Student’s t test. For multiple comparisons, statistical significance was determined using a one-way ANOVA followed by a post-hoc test (Dunn’s Multiple Comparison Test). ***, P < 0.001. Data distribution was assumed to be normal, but this was not formally tested. Scale bars: 10 μm. Colored asterisks indicate intestinal lumen. A dotted line indicates the outline of the intestine. Source data are available for this figure: SourceData F2.