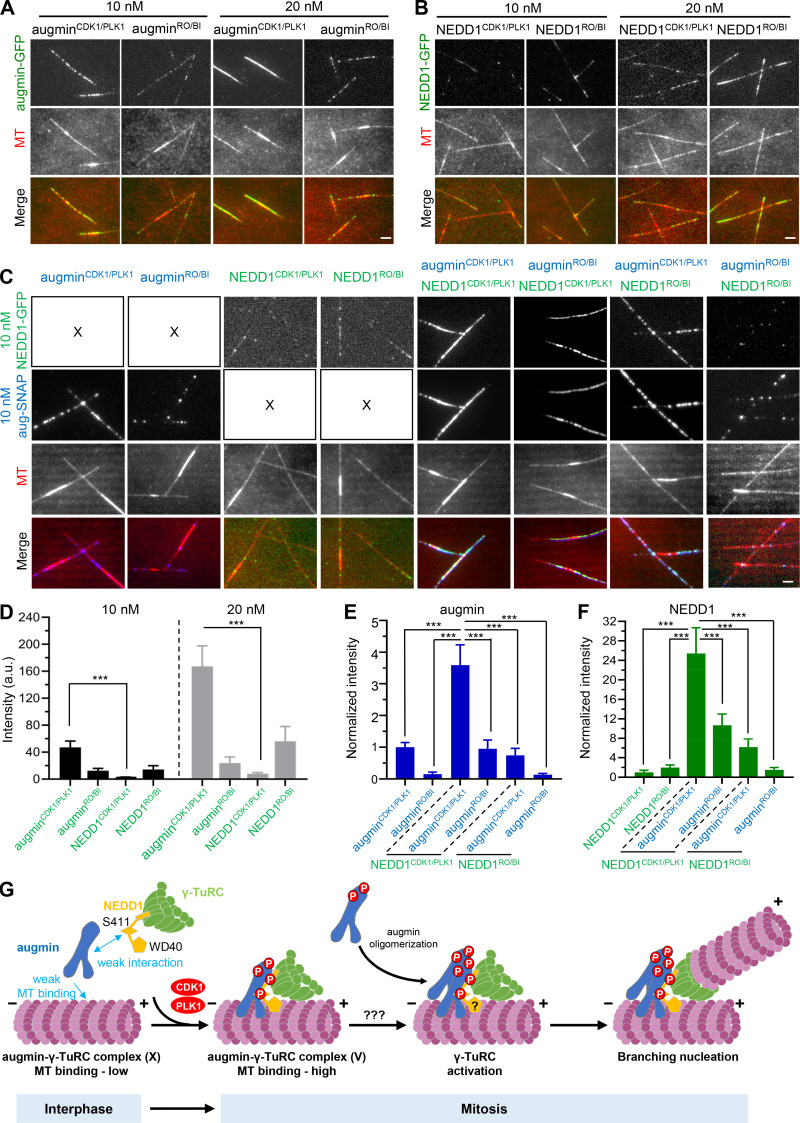
Figure 9.
Different roles for NEDD1 phosphorylation and augmin phosphorylation in their synergistic MT-binding activities. (A and B) TIRF microscopy images of MTs (red) grown in the presence of augminCDK1/PLK1-GFP, augminRO/BI-GFP, NEDD1CDK1/PLK1-GFP, or NEDD1RO/BI-GFP (green) at indicated concentrations. (C) TIRF microscopy images of MTs (red) grown in the presence of different versions of 10 nM augmin-SNAP (blue; augminCDK1/PLK1 and augminRO/BI) and 10 nM NEDD1-GFP (green; NEDD1CDK1/PLK1 and NEDD1RO/BI), either alone or combined in pairs. (D) Quantification of intensities of indicated versions of augmin and NEDD1 along MTs for the experiments shown in A and B. n = 60 MTs from three experiments. (E and F) Quantification of intensities of indicated versions of augmin-SNAP (E) and NEDD1-GFP (F) along MTs for the experiments shown in C. The values were normalized to the intensity of augminCDK1/PLK1-SNAP alone (E) or NEDD1CDK1/PLK1-GFP alone (F). n = 40 MTs from three experiments. (G) Model for cell cycle–dependent regulation of branching MT nucleation. In interphase, the interaction between augmin and γ-TuRC is weak, and the MT-binding affinity of augmin is also low. Therefore, branching MT nucleation does not occur at this stage in the human system. Upon entry into mitosis, CDK1- and PLK1-mediated phosphorylation drives the formation of a stable MT branching machinery, the complete augmin–γ-TuRC complex, and promotes a synergistic enhancement of the MT-binding activity of this machinery. NEDD1 WD40 domain plays an essential role in activating the branching nucleation via a yet unknown mechanism. Augmin oligomerization may also play an important role in the activation step. Scale bars, 2 μm. Data represent mean ± SD. ***, P < 0.001, two-tailed t test.