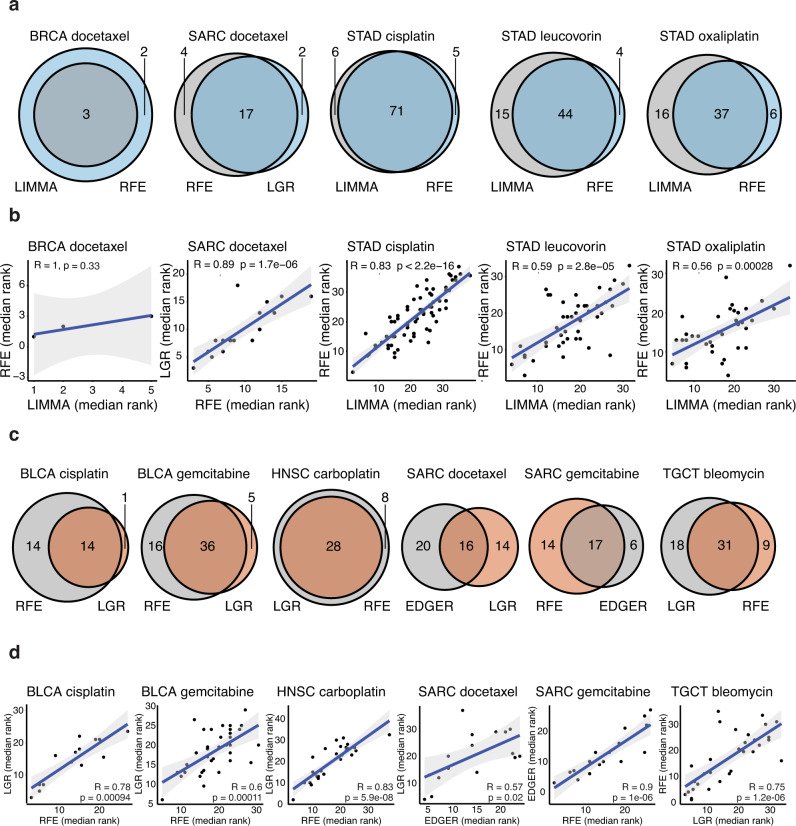
Fig. 5. Comparison of drug response model top-ranked selected features by each ML method. For each drug response model, we selected the two best ML methods by significance for the prediction improvement over their respective clinical covariate-only model.
Venn diagrams for microbial abundance (a) or gene expression (c) models comparing the number of features individually selected by each ML method, and the intersection of the two ML methods. Spearman rank correlation plots for microbial abundance (b) or gene expression (d) models showing that the median rank of features (among the 100 model instances in which the feature was selected) often correlated between the two most significant ML methods; p-values are two-sided. The best method is shown on the x-axis, the second best on the y-axis. Source data are provided as a Source Data file. The number of cases involved in each experiment are shown in Supplementary Table 1.