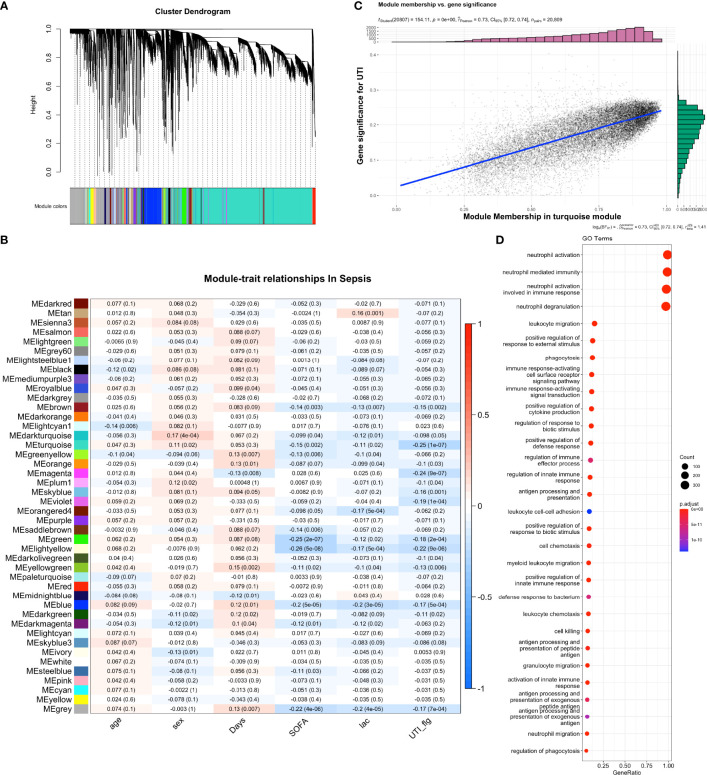
Figure 3.
Weighted gene co-expression network analysis (WGCNA) of bulk RNA-seq samples. (A) Dendrogram clustering of the co-expressed genes into modules. Modules were labeled by different colors. (B) module-trait relationship in sepsis samples. The blue color indicates negative correlation and the red color indicates the positive correlation. The numbers in each cell are Pearson’s correlation coefficients between module eigengene and trait values, with p values shown in the parenthesis. (C) Correlation between module membership and gene significance for UTI. Each dot represents a gene. Gene significance was quantified by comparing gene expression between UTI and control groups. Module membership of a given gene was quantified by correlating its expression profile with the module eigengene. (D) Gene set enrichment analysis of the turquoise module with GO terms.