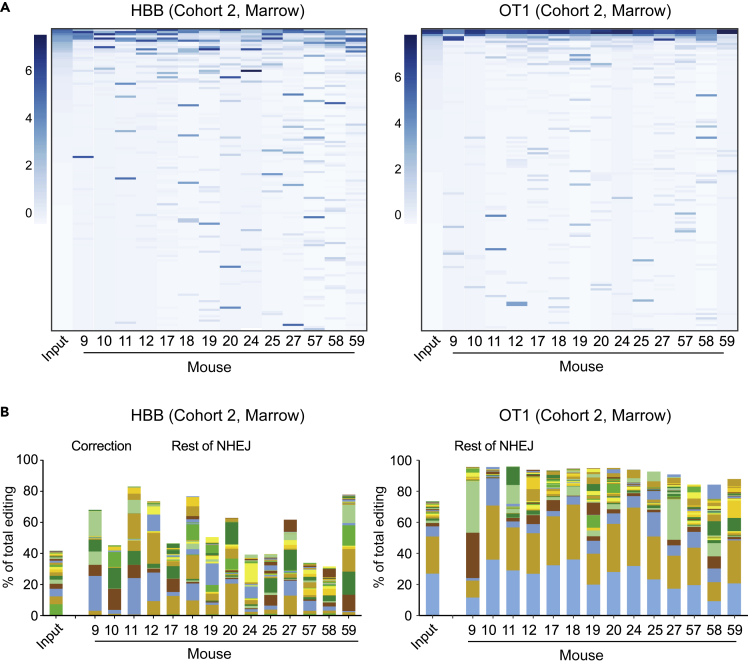
Figure 3.
Analysis of allelic diversity in edited and xenografted hematopoietic cells
(A) Heatmaps of abundance of alleles (rows) in input sample (first column) and xenografted marrow of Cohort two mice. Alleles are sorted (vertically) by decreasing abundance in the input sample, and filtered for alleles with at least 0.1% abundance in some mouse or the input, resulting in 140 HBB and 135 OT1 alleles. The scale at the left of each heatmap indicates the band intensity corresponding to the scaled log2 abundance (i.e., intensity corresponding to four indicates that the band made up 16% of reads in that marrow). Note that the most common alleles in the input population are of minor abundance in some mice, while alleles that are rare in the input may be common in one or more mice. Heatmaps for Cohorts 1, 3, and four are Figure S3.
(B) Stacked bar graphs indicating the contribution of the top 24 indel alleles (solid bars) at HBB (left) and OT1 (right) in each Cohort two mouse, as a fraction of total edited alleles. The remainder of NHEJ alleles are contained in brown striped bars (top segment); the corrected HBB alleles (HDR) are shown as a blue striped bar (second segment from top). Stacked bar graphs for all cohorts in a form that allows identification of the indel corresponding to each color are in the supplementary Excel workbook.