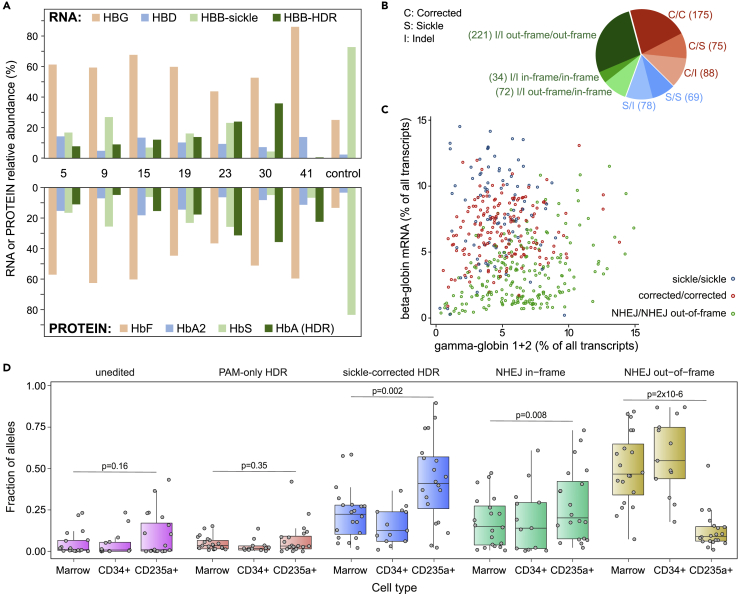
Figure 4.
Globin expression, allelic assortment, and in vivo selection for corrected HBB genotype in erythroid cells
(A) RNA-Seq (upper panel) and HPLC (lower panel) on erythrocytes differentiated in bulk liquid culture from marrow human CD34+ cells of eight xenografted mice (mouse numbers are between upper and lower panels). The control was engrafted with unedited cells. For RNA-Seq, only β-like globins are shown, in colors matching the corresponding hemoglobin assayed by HPLC. HPLC traces are shown in Figure S4.
(B) HBB Genotypes inferred from individual RNA-Seq of 812 clonal erythroid colonies differentiated from edited human CD34+ cells; colonies with two NHEJ alleles (green) are further characterized by in- or out-of-frame deletions.
(C) Relationship between the expression of γ-globin and β-globin in erythroid colonies with homozygous genotypes. Dots represent individual colonies; the xaxis represents the cumulative expression of HBG1 and HBG2; the yaxis represents expression of HBB. Expression is reported as the percentage of the expression of all transcripts within a colony. Colonies with out-of-frame indels at HBB have a reduced ratio of HBB to HBG mRNA. See also Figure S6.
(D) Distribution (boxplot) of HDR and NHEJ allele frequencies in total marrow genomic DNA, marrow human CD34+ genomic DNA, and mRNA from marrow human erythroid (CD235+) cells; data for individual mice are shown as circles. A “sickle corrected” HDR allele has both the PAM mutation and sickle correction, while a “PAM-only” HDR allele has the PAM mutation but no sickle correction. An “in-frame” NHEJ allele has an indel that maintains the β-globin reading frame, while an “out-of-frame” NHEJ allele disrupts it. The corrected allele is enriched in erythroid cells compared to marrow and CD34+ cells, while the NHEJ out-of-frame allele is severely depleted in erythroid cells; see also Figure S8. p-values are calculated using the Wilcoxon Rank-Sum test. Error bars depict standard deviation from the mean.