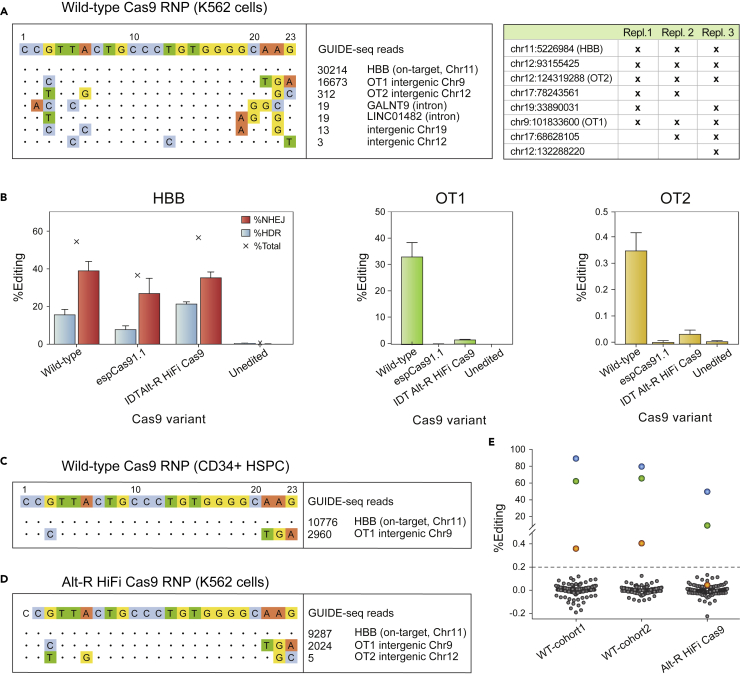
Figure 5.
Assessment of off-target cleavage by the Cas9 RNP
(A) Representative GUIDE-seq with the Cas9 RNP in K562 cells (left), and genomic coordinates of GUIDE-seq hits detected in three independent replicates (table at right).
(B) Comparison of editing at HBB by high-fidelity Cas9 variants espCas9-1.1 (Vakulskas et al., 2018) and Alt-R HiFi Cas9 (Kleinstiver et al., 2016) in healthydonor HSPCs. Error bars depict standard deviation from the mean.
(C) GUIDE-seq in CD34+ HSPCs edited with the 3xMS-G10 RNP with wild-type Cas9. Only two sites, the on-target site in HBB and the primary off-target site OT1, are detected.
(D) Representative GUIDE-seq with Alt-R HiFi Cas9 RNP in K562 cells.
(E) Total gene editing rates (%HDR + %NHEJ) measured by pooled-primer PCR at 190 of 201 identified off-targets, in the edited HSPCs injected into Cohorts one and 2 (“input” in Figure 1C), and in healthy donor HSPCs edited with Alt-R HiFi Cas9; indels observed at the same sites in untreated cells are subtracted. Blue dots: on-target HBB; green dots: OT1; orange dots: OT-II. Editing at >0.2% of alleles (dashed line) by wild-type Cas9 is observed only at HBB, OT1, and OT-II, and by high-fidelity Cas9 only at HBB and OT1.