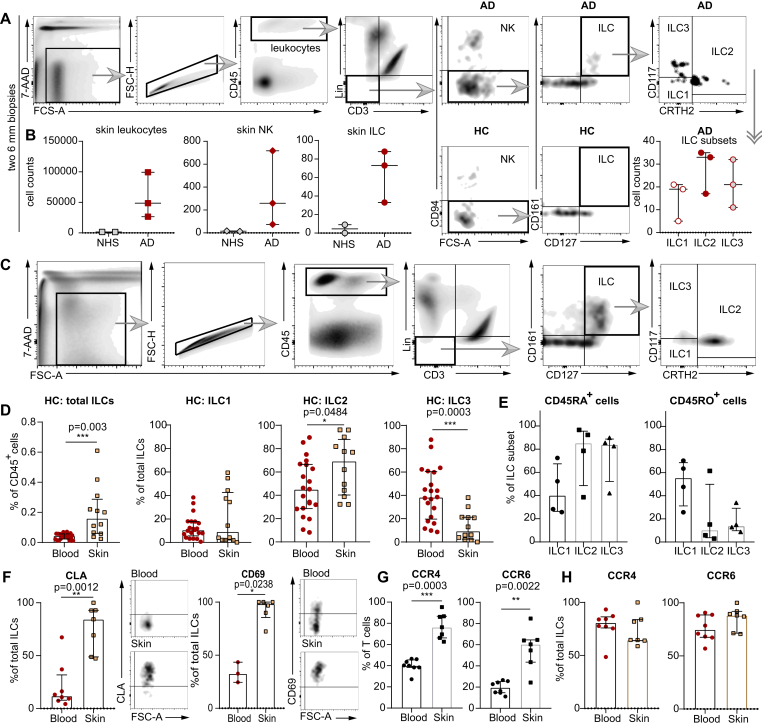
Fig 1.
Characterization of ILCs with flow cytometry. A, Gating strategy for ILCs from skin punch biopsies. B, Numeric differences in leukocytes, NK cells, and ILCs between NHS (n = 2) and AD skin (n = 3) as well as ILC subpopulations detectable in AD skin samples (Lin for [A] and [B] without CD94 and CD16). C, Gating strategy for ILCs and ILC subsets extracted from NHS sheets and PB. D, Total ILCs and ILC subsets from NHS sheets (n = 12) and PB of HCs (n = 21). E, CD45RA+ and CD45RO+ ILC subsets in NHS (n = 4). F, Expression of CLA and CD69 on ILCs from NHS and PB of HCs (NHS [n = 7] and PB [n = 8] for CLA; NHS [n = 6] and PB [n = 3] for CD69). CD45+CD3+ T cells (G) and ILCs (H) expressing CCR4 and CCR6 in NHS (n = 7) and PB (n = 8). Dot plots represent medians with interquartile ranges (IQRs). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. 7-AAD, 7-Aminoactinomycin D, FSC-A/H, forward scatter area/height; Lin, lineage cocktail (TCRα/β, TCRγ/δ, CD19, CD16, CD15, CD14, CD94, CD11c, BDCA-2, FcεRIα, glycophorin A, CD41, CD1a, and CD34).