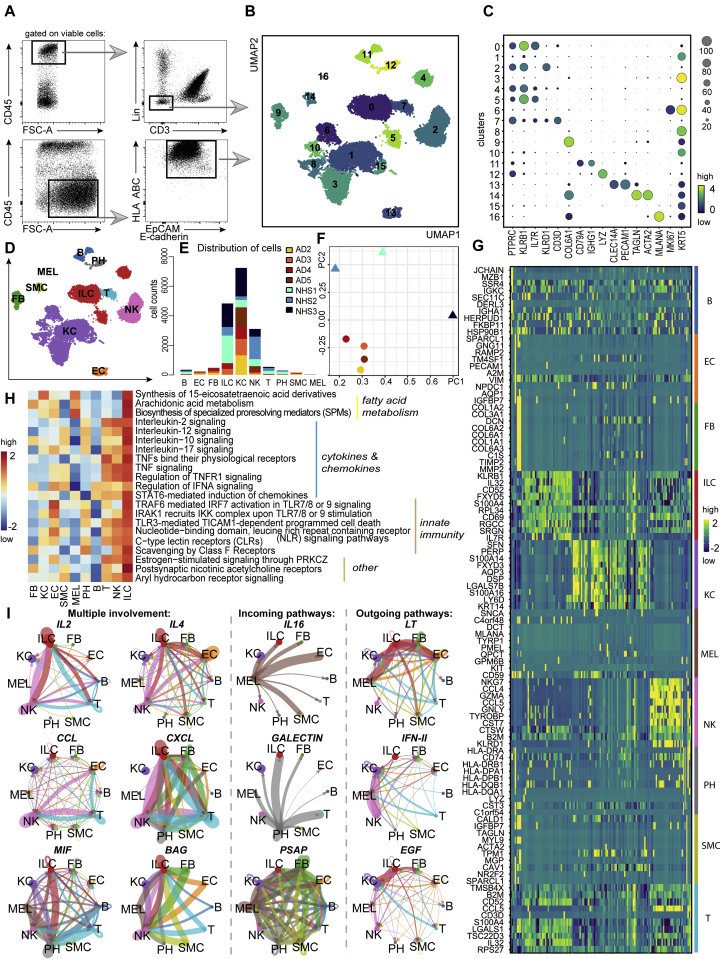
Fig 3.
Characterization of RNA-sequenced cutaneous ILC-enriched samples. A, Gating strategy of ILC-enriched samples sorted for scRNA-seq as viable CD45+Lin–CD3– cells (NHS, n = 3; AD skin, n = 4) and supplemented with CD45–EpCAM/E-cadherin+HLA ABC+ keratinocytes (NHS, n = 1; AD skin, n = 4) (lineage cocktail without CD94 and CD16). B, Uniform Manifold Approximation and Projection for Dimension Reduction (UMAP) plot (17,307 cells) of ILC-enriched samples sorted from NHS sheets (n = 3) and AD skin biopsies (n = 4). C, Dot plot demonstrating mean loge normalized expression of cluster-specific marker genes. Color scale indicates mean expression, and circle size indicates fraction of cells in a group. D, Cell types present in the sequenced data set (NHS, n = 3; AD skin, n = 4) from (B) and represented by using a UMAP plot. E, Numeric distribution of cells among cell types depicted in (D) and color-coded on the basis of sample origin (see Table E5 [in the Online Repository at www.jacionline.org]). F, Principal component analysis of the sequenced samples. G, Heatmap, indicating the top 10 differentially expressed genes (P < .05) between annotated cell types from (D) (see Table E3). H, Major ILC-related pathways, as determined with the ReactomeGSA package; pathway expression levels are shown as z score normalized values. I, ILC-related interaction patterns, as revealed by using the CellChat toolkit.