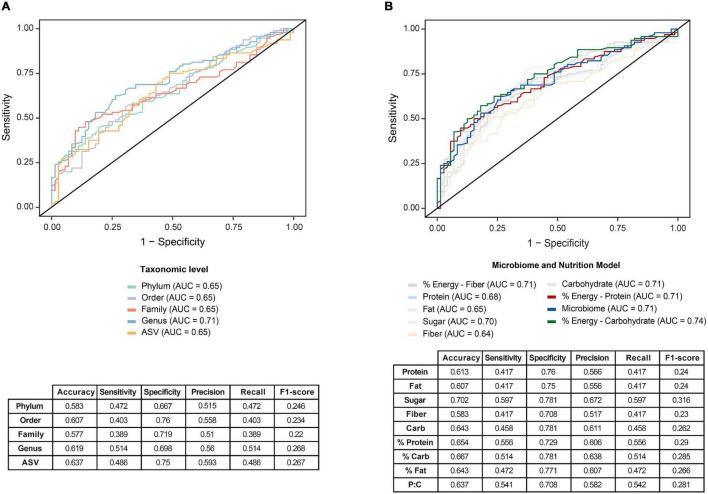
FIGURE 4.
Predictive modeling to identify Parkinson’s disease (PD) was optimized by a two-stage model that incorporates nutritional and microbiome data. (A) Predictive Random Forest modeling was undertaken to identify the utility of the gut microbiome as a potential signature for PD. Comparisons at five taxonomic levels; phylum, order, family, genus and ASV, were used to predict PD, with greatest predictive capacity provided at the genus level on receiver operating characteristic curve (ROC), with an area under the curve (AUC) of 0.71. (B) An optimized two-stage predictive Random Forest model analysis was subsequently undertaken that considered dietary intake as an influence on the gut microbiome. Comparing the utility of the one-stage microbiome model (AUC = 0.71) with the two stage model, a slightly improved predictability was achieved. when incorporating dietary macronutrient data. Specifically, the incorporation of carbohydrate contribution to total energy in the model improved the prediction (AUC = 0.74), whereas incorporation of fiber, fat or protein macronutrient data alone into the model did not improve the predictive potential to identify PD. Accompanying sensitivity and specificity analyses are presented in the tables.