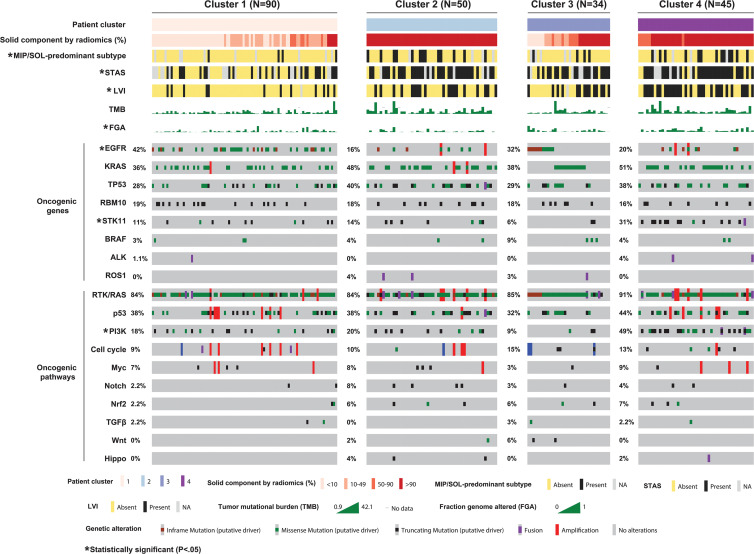
Figure 3:
OncoPrint of clinical-pathologic and genomic variables for all patients with clinical stage I lung adenocarcinoma broken down by radiomic consensus clustering. Columns represent patients within each cluster of radiomics features, and rows represent the frequencies of alterations of all individually analyzed oncogenic genes and 10 canonical oncogenic pathways. Cluster characteristics were compared using Fisher or χ2 exact test for categorical data and Kruskal-Wallis test for continuous data. * Associations between radiomic clusters and clinicopathologic or genomic variables are significant (P < .05). FGA = fraction of genome altered, LVI = lymphovascular invasion, NA = not applicable, MIP = micropapillary, SOL = solid, STAS = spread through air spaces, TMB = tumor mutational burden.