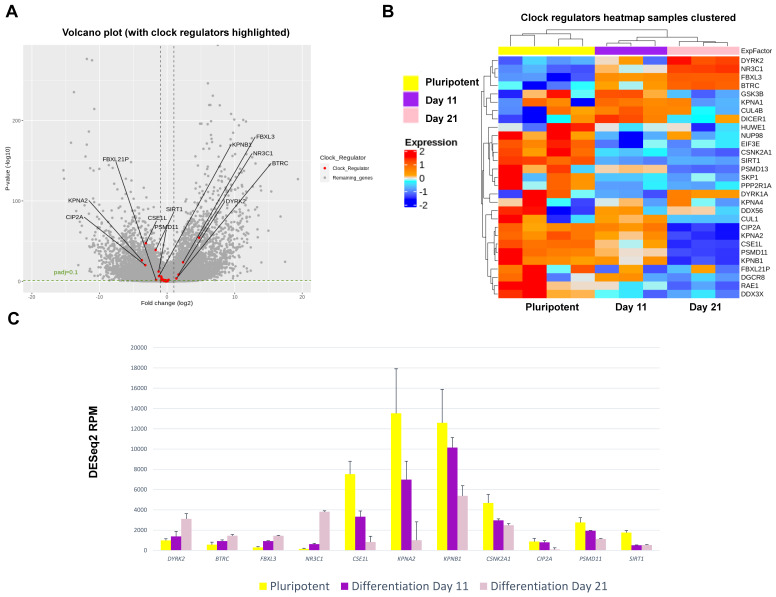
Figure 8.
Expression changes for clock regulator genes as revealed by RNAseq during chondrogenic differentiation of MAN13 cells. (A). Volcano plot of differential expression analysis showing all differentially expressed genes during the chondrogenic developmental stages. Data were plotted against -Log10 p-value. Horizontal dotted lines represent significance level of padj < 0.1. Vertical dotted lines indicate Log2 fold change of -1 and 1 between Day 21 vs. Day 11. Significant clock regulator genes with padj < 0.1 and log2FC > 1 or log2FC < -1 were highlighted in red with gene names. (B). Heatmap of the clock regulator genes that showed significant variations among the three time points, as revealed by differential expression analysis. (C). RNAseq data (DESeq2 normalised counts) for clock regulator genes that showed most significant variations in expression levels among the three time points (padj < 0.001). Between Day 21 and Day 11: padj < 0.05 for CSNK2A1; not significant for SIRT1; padj < 0.001 for all the other genes.