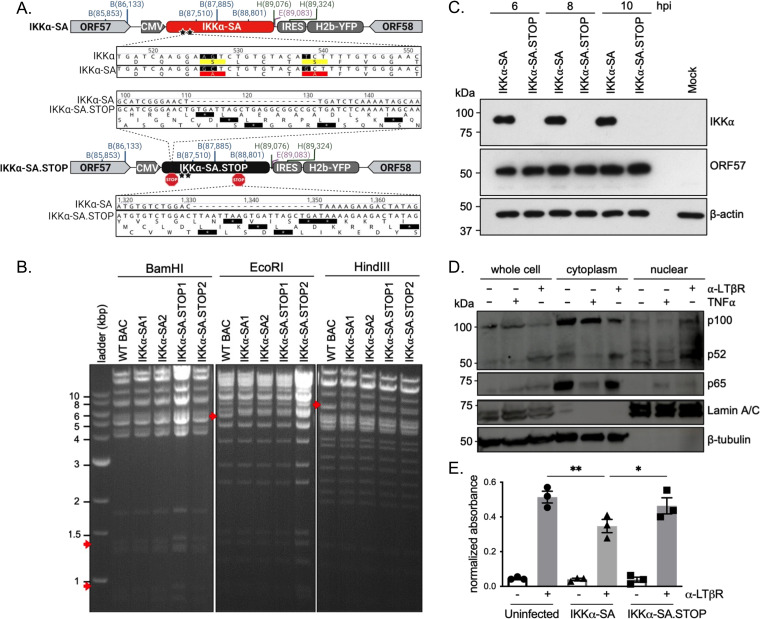
FIG 1.
Generation of recombinant MHV68 expressing a transgene that impairs IKKα signaling. (A) Schematic of IKKα-SA and IKKα-SA.STOP viruses, made with BioRender. A cassette encoding a CMV-IE promoter driving IKKα-SA or IKKα-SA.STOP, an IRES, and H2B-YFP was inserted into the neutral locus between ORF57 and ORF58 using BAC-mediated recombination. Stars denote the locations of S176A and S180A mutations; STOP indicates an all-frame stop cassette insertion. (B) In the BamHI digest, IKKα-SA virus has the appearance of two bands at 920 bp and 1,381 bp, and IKKα-SA.STOP virus has the appearance of two bands at 940 bp and 1,402 bp (red arrows), all compared to the parental WT BAC. The EcoRI digest reveals the appearance of a 5,438-bp band and a 5,615-bp band in both IKKα-SA and IKKα-SA.STOP viruses compared to the parental WT BAC. Finally, in the HindIII digest, there is the appearance of a 6,570-bp band in both IKKα-SA and IKKα-SA.STOP compared to the parental WT BAC. (C) Primary WT MEFs were infected with IKKα-SA or IKKα-SA.STOP at an MOI of 10. Immunoblots of protein lysates were analyzed at 6, 8, and 10 hpi for the IKKα transgene and the IE viral protein ORF57. β-Actin was used as a loading control. (D) Primary WT MEFs were stimulated with 1 μg/mL anti-LTβR for 18 h or 50 ng/mL TNF-α for 15 min. Cell lysates were fractionated to collect cytoplasmic and nuclear fractions. p100 cleavage and p65 were detected by immunoblotting. Lamin A/C was the control for the nuclear fraction; β-tubulin was the control for the cytoplasmic fraction. (E) Primary MEFs were infected with MHV68-IKKα-SA or -IKKα-SA.STOP at an MOI of 10. At 4 hpi, cells were stimulated with 0.3 μg/mL of anti-LTβR antibody for 5 h. At 9 hpi, cells were harvested. Nuclear fractions were probed for p52 activation using a p52 ELISA and normalized to uninfected, unstimulated cells. Results represent means ± standard errors of the means (SEM) for three independent experiments with duplicate samples per replicate. ***, P < 0.001; ****, P < 0.0001 (analyzed using ANOVA with Sidak’s multiple-comparison posttest).