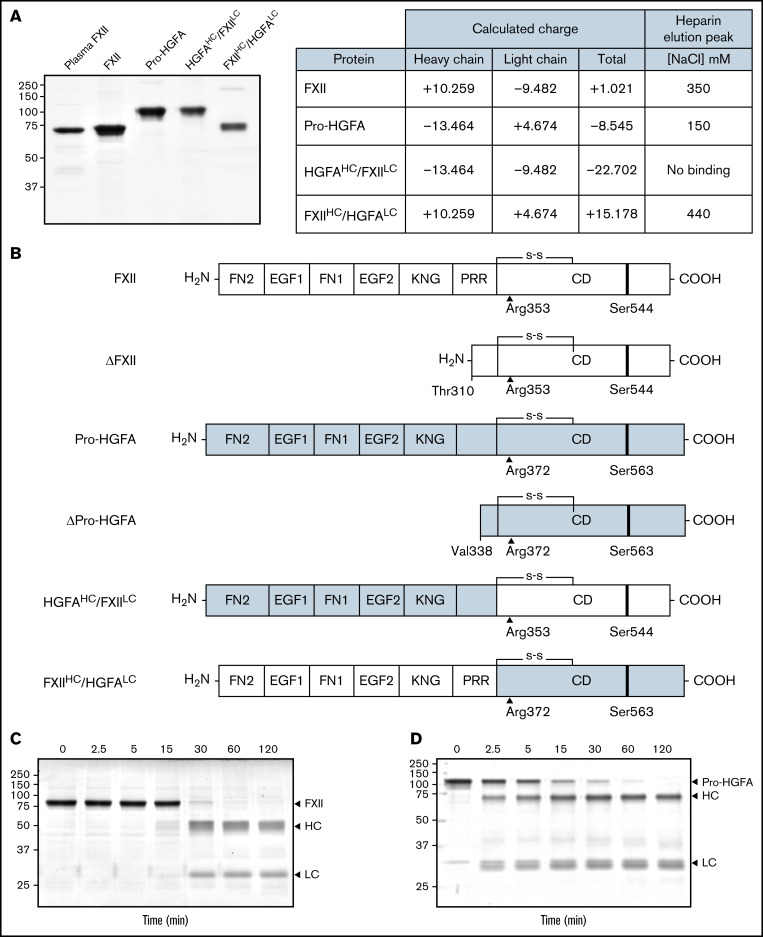
Figure 1.
Recombinant FXII, Pro-HGFA, and heavy chain/light chain chimeras. (A) Nonreducing sodium dodecyl sulfate–polyacrylamide gel electrophoresis of purified plasma-derived FXII and recombinant wild-type FXII (FXII), wild-type Pro-HGFA, and chimeras HGFAHC/FXIILC and FXIIHC/HGFALC (2 μg per lane). Positions of molecular mass standards in kilodaltons are shown to the left of the image. The table on the right shows predicted net charges on the proteins, with separately determined total, heavy chain, and light chain values. The right column of the table indicates the NaCl concentration (mM) required to elute each protein off of a heparin-sepharose column. (B) Schematic diagrams of the structures of the proteins shown in panel A. FXII domains are indicated in white, and Pro-HGFA domains in gray. The noncatalytic domains of FXII are FN2, EGF1, FN1, EGF2, and KNG domains and a PRR. Pro-HGFA is organized similarly except that it does not have a PRR, and the corresponding sequence has not been assigned a name. Positions of FXII and Pro-HGFA active site serine residues (Ser544 and Ser563, respectively) are indicated by black bars, and sites for proteolytic needed activation (after Arg353 and Arg372, respectively) are indicated by black arrows. Also shown are the truncated forms ΔFXII and ΔPro-HGFA. ΔFXII is formed by cleavage of FXII after residue 309, while cleavage of Pro-HGFA after Arg337 forms ΔPro-HGFA. (C-D) Time courses of recombinant FXII (200 nM) incubated with poly-P (70 μM) (C) and Pro-HGFA incubated with thrombin (26 nM) and dextran sulfate (10 μg/mL) (D). Positions of standards for zymogen (Z) FXII and Pro-HGFA and the heavy and light chains of FXIIa and HGFA are indicated at the right of each image. Positions of molecular mass standards in kilodaltons are shown to the left of the images.