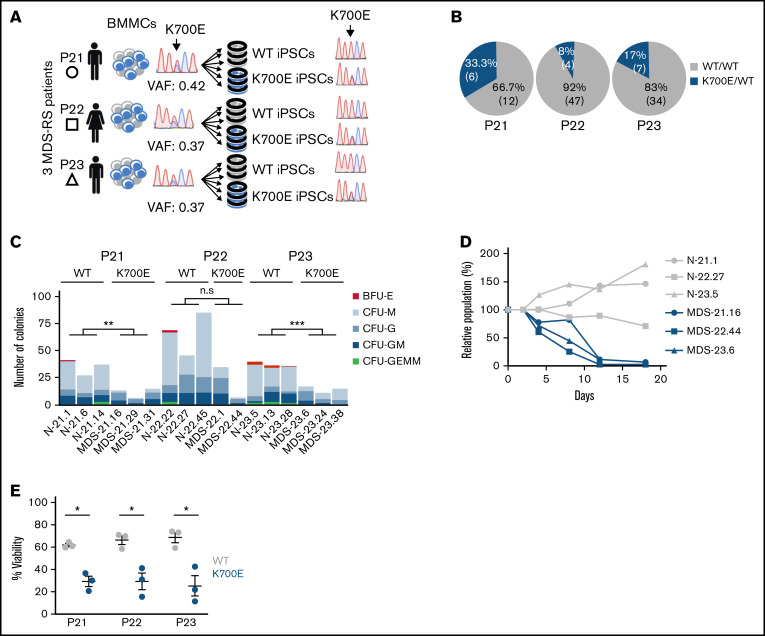
Figure 1.
Derivation and phenotypic characterization of MDS-RS patient-derived SF3B1K700E and SF3B1WT iPSCs. (A) Schematic overview of the derivation of iPSC lines with isolated SF3B1K700E mutation and genetically matched normal WT lines from 3 patients with MDS-RS (BMMCs, bone marrow mononuclear cells). (B) Percentage and absolute number of iPSC colonies of each genotype derived from each patient sample. (C) Methylcellulose assays on day 14 of hematopoietic differentiation. The number of colonies from 5000 seeded cells is shown per iPSC line (CFU-GEMM, colony-forming unit-granulocyte, erythrocyte, monocyte, megakaryocyte; CFU-GM, CFU-granulocyte, monocyte; CFU-G, colony-forming unit-granulocyte; CFU-M, CFU-monocyte; BFU-E, burst-forming unit-erythrocyte); n.s., not significant; **P ≤ .01; ***P ≤ .001. (D) Competitive growth assay. The cells were mixed 1:1.5 at the onset of hematopoietic differentiation with a genetically matched WT iPSC line stably expressing green fluorescent protein (GFP). The relative population size was estimated as the percentage of GFP− cells (measured by flow cytometry) at each time point (days 4-18 of differentiation), relative to the percentage of GFP− cells on day 2. (E) Percent viable (4′,6-diamidino-2-phenylindole−) cells on day 14 of hematopoietic differentiation of the indicated SF3B1K700E and SF3B1WT iPSC-HSPCs. Mean and standard error of the mean (SEM) of 1 to 3 independent differentiation experiments with 2 to 3 iPSC lines from each group are shown. *P ≤ .05.