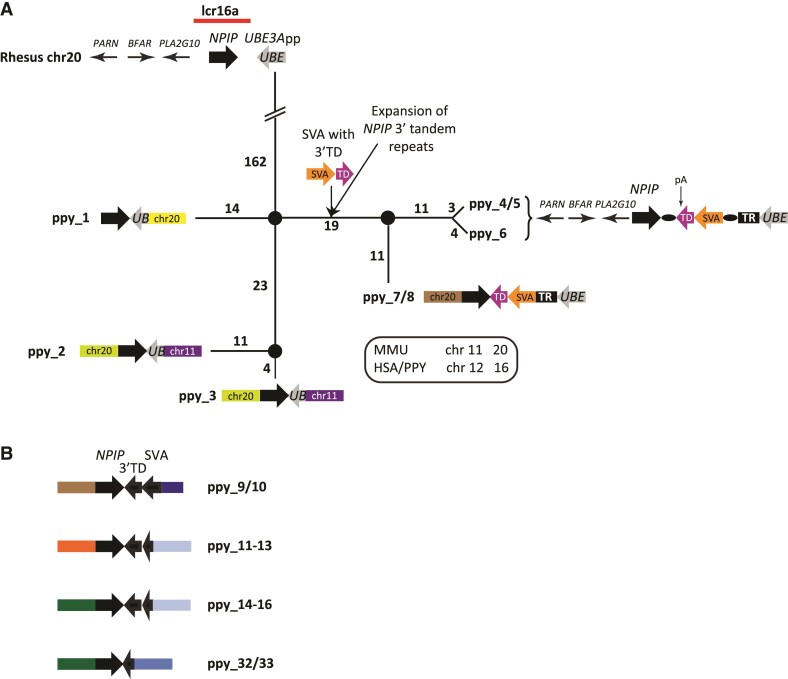
Fig. 2.
SVA is an integral component of lcr16a in orang-utan. (A) Median-joining network of orang-utan lcr16a copies containing the UBE3A processed pseudogene. The network is rooted on the rhesus macaque UBE3A processed pseudogene (UBE3App) which is shown in the context of the rhesus ancestral lcr16a locus on top of the network. Except for the path connecting the root and the first node the network is drawn to scale. The number of substitutions is indicated for each path. In ppy_1–3, the lcr16a 3′ breakpoint localizes to UBE3App (UBE—gray arrows). Copies ppy_4–8 are characterized by the insertion of an SVA carrying a 3′ TD into the expanded NPIP 3′ tandem repeats. TSDs (black ovals) are discernible in ppy_4–6 which, based on the order of the upstream genes, represent the orang-utan lcr16a ancestral locus. TR—tandem repeats downstream of the SVA integration site; pA—potential polyadenylation signal provided by the SVA 3′ transduction. Chromosome numbers refer to the rhesus genome; a key with the respective numbers in human and orang-utan is provided. MMU, Macaca mulatta; HSA, Homo sapiens; PPY, P. (pygmaeus) abelii. (B) Schematic representation of additional lcr16a copies present on orang-utan chromosomes 16 and 11 (ppy_33). Note that in all cases, the 3′ breakpoint localizes to either the SVA or its 3′ transduction. Colored boxes in (A) and (B) represent lcr16a flanking sequences or flanking SDs (not drawn to scale). Identical colors indicate identical mapping positions (rheMac10) and junctions between the modules and the low copy repeat. The key to the color code is provided in supplementary table S2, Supplementary Material online. For reasons of clarity only immediately flanking modules are shown.