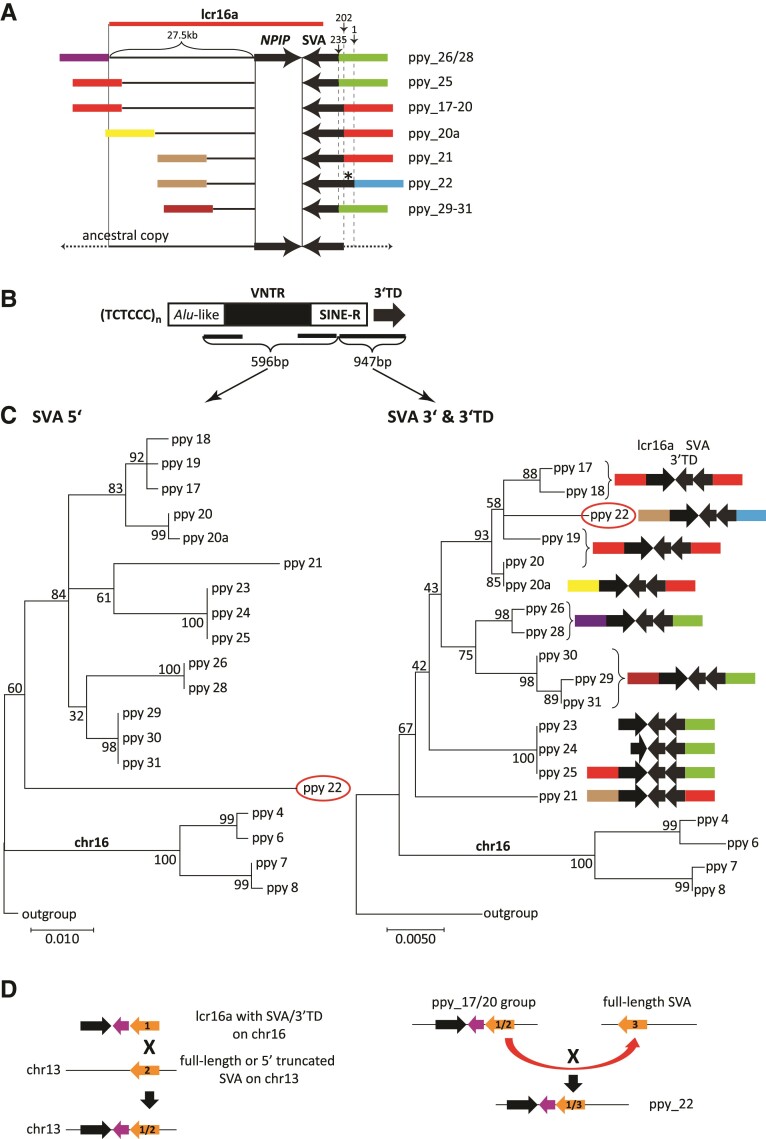
Fig. 3.
SVA recombination contributes to lcr16a interchromosomal and intrachromosomal mobility in orang-utan. (A) Structure and extension of the lcr16a core duplicon in duplication blocks on chromosome 13. Only immediately adjacent flanking sequences/SDs (colored boxes) are shown. Thin horizontal lines denote lcr16a sequence upstream of the NPIP coding region (drawn to scale). Numbers above vertical arrows refer to the SVA 5′ end relative to the SVA_A RepeatMasker consensus sequence. Ppy_22 carries a full-length SVA (asterisk) likely resulting from SVA inter-element recombination. Minimal 5′ and 3′ extension of the hypothetical chromosome 13 ancestral lcr16a copy is shown at the bottom. Note that the red boxes at the 5′ and 3′ ends, respectively, represent modules derived from the same locus (separated by 1.1 kb). (B) Schematic representation of an SVA element. TCTCCC hexameric repeats at the 5′ end are followed by an Alu-like domain, a VNTR and a retrovirus-derived SINE-R. The element present in orang-utan lcr16a carries a 3′ TD. Black bars indicate the regions used in the phylogenetic analysis shown in C. Numbers denote the length of the sequences used for the analysis. (C) Different phylogenetic relationships of SVA 5′ and 3′ ends suggest recombination as the mechanism of lcr16a interchromosomal (chromosome 16 to chromosome 13) and intrachromosomal (ppy_22 on chromosome 13) duplicative transposition. Maximum likelihood phylogenetic trees (Kimura two-parameter; n = 10,000 bootstrap replicates) were generated for SVA sequences up- and downstream of the potential recombination region (for details see main text and supplementary fig. S1, Supplementary Material online). The SVA 5′ region comprises the part of the Alu-like domain present in all copies, the first and the last three VNTR subunits as well as the 5′ part of the SINE-R. The central part of the VNTR could not be aligned. The downstream part spans the SINE-R 3′ and the 3′ transduction. Low bootstrap values of, in particular, the interior nodes can be attributed to the relatively short length of the sequences used in the analysis. (D) Schematic representation of SVA recombination in interchromosomal (left panel) and intrachromosomal (right panel) lcr16a duplicative transposition. SVAs are shown in orange; the 3′ transduction in purple. The black arrow denotes lcr16a. Numbers indicate the different SVA elements involved in recombination. X denotes recombination events. Colored boxes in (A) and (C) represent lcr16a flanking sequences or flanking SDs (not drawn to scale). Identical colors indicate identical mapping positions and junctions between the modules and lcr16a at its 5′ and 3′ end, respectively. Modules derived from the same locus are presented in the same color. The key to the color code is provided in supplementary table S3, Supplementary Material online. For reasons of clarity only immediately flanking modules are shown.