Abstract
NDST1 (glucosaminyl N-deacetylase/N-sulfotransferase) is a key enzyme in heparan sulfate (HS) biosynthesis, where it is responsible for HS N-deacetylation and N-sulfation. In addition to the full length human enzyme of 882 amino acids, here designated NDST1A, a shorter form containing 825 amino acids (NDST1B) is synthesized after alternative splicing of the NDST1 mRNA. NDST1B is mostly expressed at a low level, but increased amounts are seen in several types of cancer where it is associated with shorter survival. In this study, we aimed at characterizing the enzymatic properties of NDST1B and its effect on HS biosynthesis. Purified recombinant NDST1B lacked both N-deacetylase and N-sulfotransferase activities. Interestingly, HEK293 cells overexpressing NDST1B synthesized HS with reduced sulfation and altered domain structure. Fluorescence resonance energy transfer-microscopy demonstrated that both NDST1A and NDST1B had the capacity to interact with the HS copolymerase subunits EXT1 and EXT2 and also to form NDST1A/NDST1B dimers. Since lysates from cells overexpressing NDST1B contained less NDST enzyme activity than control cells, we suggest that NDST1B works in a dominant negative manner, tentatively by replacing the active endogenous NDST1 in the enzyme complexes taking part in biosynthesis.
Keywords: alternative splicing, golgi enzyme, heparan sulfate biosynthesis, NDST
Introduction
Heparan sulfate (HS) proteoglycans are synthesized by the vast majority of cells in the human body (Sarrazin et al. 2011; Iozzo and Schaefer 2015; Lindahl et al. 2015). In the order of 20 different proteins have been identified as HS proteoglycan core proteins, each carrying a few covalently bound HS side chains. HS proteoglycans localize primarily to the cell surface but are also found in basement membranes (Iozzo and Schaefer 2015; Lindahl et al. 2015). HS is essential for embryonic development; mouse embryos lacking HS die before gastrulation starts (Lin et al. 2000). Besides participating in the generation and stabilization of morphogen gradients in the developing embryo, a major function of the HS proteoglycans is to act as coreceptors for growth factors and cytokines (Kjellen and Lindahl 2018). The HS sulfation patterns, characteristic for each cell type at a given time point, will modulate signaling as well as migration and differentiation (Bulow and Hobert 2006). In the adult human being, alterations in HS sulfation are associated with cancer but also with other pathologies including amyloid diseases, neurological disorders, infectious diseases, and inflammatory conditions (Lindahl and Kjellen 2013).
HS biosynthesis depends on a complex machinery of enzymes including multiple glycosyltransferases, sulfotransferases, and an epimerase, all residing in the Golgi cisternae (Kreuger and Kjellen 2012). After biosynthesis of the linkage tetrasaccharide sequence (glucuronic acid-galactose-galactose-xylose), HS chain formation is initiated by the EXTL3 glycosyltransferase that adds an N-acetylglucosamine (GlcNAc) residue to the linker tetrasaccharide and polymerization is started. EXT1 and EXT2, which together form the functional HS–polymerase complex (Busse-Wicher et al. 2014), then transfer alternating units of glucuronic acid (GlcA) and GlcNAc to the growing chain. Modification of the HS backbone begins already during polymerization with N-deacetylation/N-sulfation of a subset of the GlcNAc residues by N-deacetylase/N-sulfotransferase (NDST) enzymes, resulting in the formation of N-sulfoglucosamine (GlcNS) residues. During this process, the acetyl group of the GlcNAc residue is removed by the N-deacetylase activity of the NDSTs to form an N-unsubstituted glucosamine (GlcNH2), which is subsequently sulfated by the N-sulfotransferase domain, using 3′-phosphoadenosine 5′-phosphosulfate (PAPS) as sulfate donor. Many of the enzymes acting after this step require GlcNS residues for substrate recognition and N-deacetylation/N-sulfation is therefore of key importance for HS overall modification (Kreuger and Kjellen 2012). The next step is epimerization of GlcA units into iduronic acid (IdoA), which is performed by a single C5 epimerase. Finally, 2-O-sulfation of a majority of the IdoA units is carried out by a 2-O-sulfotransferase followed by glucosamine 6-O-sulfation and 3-O-sulfation. The HS biosynthesis product typically displays highly sulfated domains (so-called NS-domains) interspersed with less sulfated or nonsulfated stretches. The length of the NS-domains as well as their distribution will affect the capacity of HS to modulate signaling (Kjellen and Lindahl 2018).
There are 4 isoforms of NDST in mammals (NDST1-4). Both N-deacetylase and N-sulfotransferase activities are present in all NDST-isoforms, but the isoforms differ significantly in their enzymatic activities (Aikawa et al. 2001); NDST2 is more potent on N-deacetylation than NDST1, while NDST1 is a better N-sulfotransferase (Pikas et al. 2000; Ringvall and Kjellen 2010). NDST3 shows N-deacetylase activity but only weak N-sulfotransferase activity, while the opposite is true for NDST4 (Aikawa et al. 2001). In a study by Deligny et al. (2016), it was demonstrated that the amount of HS was increased in HEK293 cells overexpressing murine NDST2 but not in cells overexpressing NDST1, suggesting that NDSTs have further regulatory functions that may not be directly linked to their enzymatic activities. This may also be the case for HS biosynthesis enzymes shown to reside outside the Golgi compartment as shown for the 3-O-sulfotransferase HS3ST2, but also for EXTL3 (Delos et al. 2018 and references therein).
NDST1 and NDST2 mRNA are expressed in all embryonic and adult tissues, albeit at different levels, while NDST3 and NDST4 mRNA are mainly expressed during embryonic development (Grobe et al. 2002). NDST1 deficient mice die shortly after birth and display respiratory failure, skeletal abnormalities, and brain defects (Aikawa et al. 2001; Grobe et al. 2002; Ringvall and Kjellen 2010). NDST1 mutations have been linked to pathology. In 2014, it was reported that missense NDST1 mutations may result in a disorder characterized by intellectual disability, muscular hypotonia, epilepsy, and postnatal growth deficiency (Reuter et al. 2014). A Canadian group later described a patient with one mutation in the N-deacetylase and one in the N-sulfotransferase domain with developmental delays, seizure, ataxia, and poor postnatal growth (Ai et al. 2006). They named the condition “NDST1 syndrome.” Recently Macchi et al. (2020) reported that NDST1 expression positively correlated with remyelination potential in multiple sclerosis patients, while Lencz et al. (2013) discovered that point mutations in the regulatory region of NDST3 are associated with schizophrenia and bipolar disorder. These results suggest that the family of NDSTs are important for neural circuit development and brain functions. In a genome-wide association study, NDST4 has also been identified as a candidate tumor suppressor gene and to be necessary for normal differentiation of colonic epithelial cell in mice (Jao et al. 2016). In this report, we describe enzymatic properties and effect on HS biosynthesis of a shorter form of NDST1, product of an alternatively spliced mRNA.
Results
While the main product of the human NDST1 gene is a protein containing 882 amino acids (here designated NDST1A), a shorter variant of 825 amino acids (NDST1B) is transcribed from an alternatively spliced mRNA which lacks exon 12 encoding the 57 amino acids starting with 716H and ending with Q772 (see Fig. 1A). To compare their enzymatic properties, HEK 293 cells stably expressing NDST1A or NDST1B, both with a C-terminal Myc/Flag-tag were successfully generated and protein expression was confirmed by western blotting (Fig. 1B). NDST1A has an apparent molecular mass of 101 kDa, while NDST1B is smaller (94 kDa).
Fig. 1.
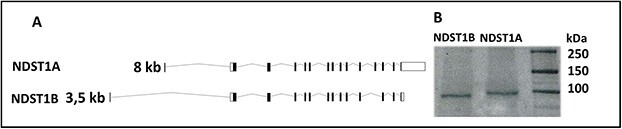
Transcripts of NDST1 and their corresponding proteins. (A) The main 8 kb transcript of the human NDST1 gene encoding the NDST1A protein containing 882 amino acids (top) and the alternatively spliced 3.5 kb transcript encoding the 825 amino acid NDST1B protein lacking exon 12. (B) SDS-PAGE followed by western blotting of cell lysates corresponding to 40 μg protein from HEK 293 cells stably overexpressing NDST1B and NDST1A. The proteins were detected with a polyclonal rabbit antibody raised against mouse NDST1. Note the size difference between NDST1A and NDST1B.
Enzyme activities of NDST1A and NDST1B cell lysates
Three different assays were applied to test the enzyme activities of the transfected cells (see Fig. 2). The N-deacetylase activity was measured with an ELISA assay where the free amino groups generated after GlcNAc N-deacetylation are recognized by a monoclonal antibody (van den Born et al. 2003); the N-sulfotransferase assay uses incorporation of 35S-sulfate from [35S]PAPS into a deacetylated polysaccharide substrate while the third assay requires both N-deacetylation and N-sulfation and measures incorporation of 35S-sulfate into a -(GlcA-GlcNAc)n- substrate. Since HEK 293 cells synthesize HS they also show a low basal level of NDST enzyme activity. HEK 293 cells transfected with an empty vector were therefore used as a control. As shown in Figure 3A, extracts of both NDST1A and NDST1B cells display N-deacetylase activity. However, while the N-deacetylase activity in lysates of NDST1A overexpressing cells is very high, NDST1B lysates contain less N-deacetylase activity than the control HEK 293 cells. Similar results were found for the N-sulfotransferase activities (Fig. 3B) as well as in the assay measuring combined N-deacetylase and N-sulfotransferase activities (Fig. 3C). Extracts of control HEK 293 cells contained roughly twice as much enzyme activity as extracts from cells overexpressing NDST1B.
Fig. 2.
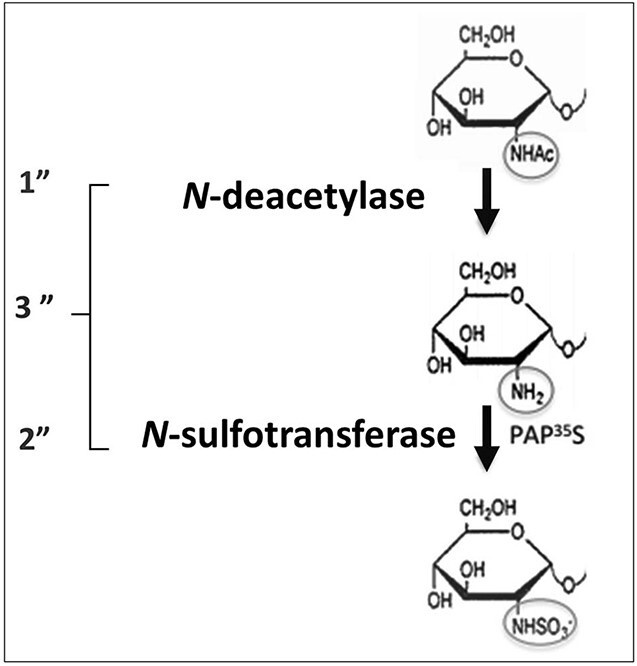
The 3 enzyme assays used to detect the 2 enzyme activities of NDST enzymes. 1” N-deacetylase activity is measured in an ELISA assay where the monoclonal antibody JM403 recognizes the free amino-groups generated after N-deacetylation. 2” N-sulfotransferase activity is measured after incorporation of 35S-sulfate groups from [35S]PAPS into the deacetylated K5-polysaccharide substrate. 3” combined N-deacetylase and N-sulfotransferase activities are measured after incorporation of 35S-sulfate groups from [35S]PAPS into the K5-substrate.
Fig. 3.
Enzyme assays of cell lysates from control HEK 293 cells and HEK 293 cells overexpressing NDST1A or NDST1B. The assays were carried out on similar volumes of lysates of stable cell lines expressing NDST1A (A1, A2) and NDST1B (B1 and B2). The values given have been corrected for protein content of the lysates. (A) N-deacetylase ELISA assay of serial dilutions of cell lysates prepared from HEK 293 cells transfected with empty vector (•), or from cells stably overexpressing NDST1A ( ) and NDST1B (
) and NDST1B ( ), respectively. (B) N-sulfotransferase activity and (C) combined N-deacetylase and N-sulfotransferase activities. The values shown are the mean of duplicates.
), respectively. (B) N-sulfotransferase activity and (C) combined N-deacetylase and N-sulfotransferase activities. The values shown are the mean of duplicates.
NDST enzyme activities of purified NDST1A and NDST1B
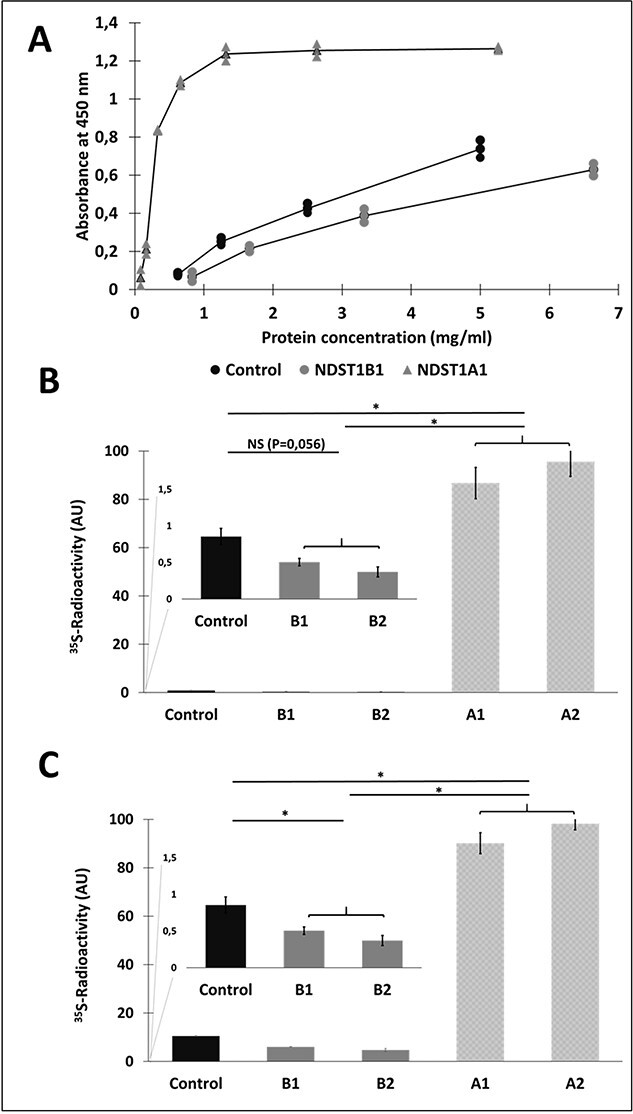
The cell lysates were used for purification of NDST1A and B using anti-Flag magnetic beads (Fig. 4A) and the purified enzymes were again assayed for enzyme activity (Fig. 4B, C, and D). HEK 293 cells transfected with empty vector were as before used as a control. However, this time endogenous NDST enzymes lacking the Flag-tag were not expected to bind to the magnetic beads. As shown in Figure 4, all 3 assays failed to detect any enzyme activity in purified NDST1B.
Fig. 4.
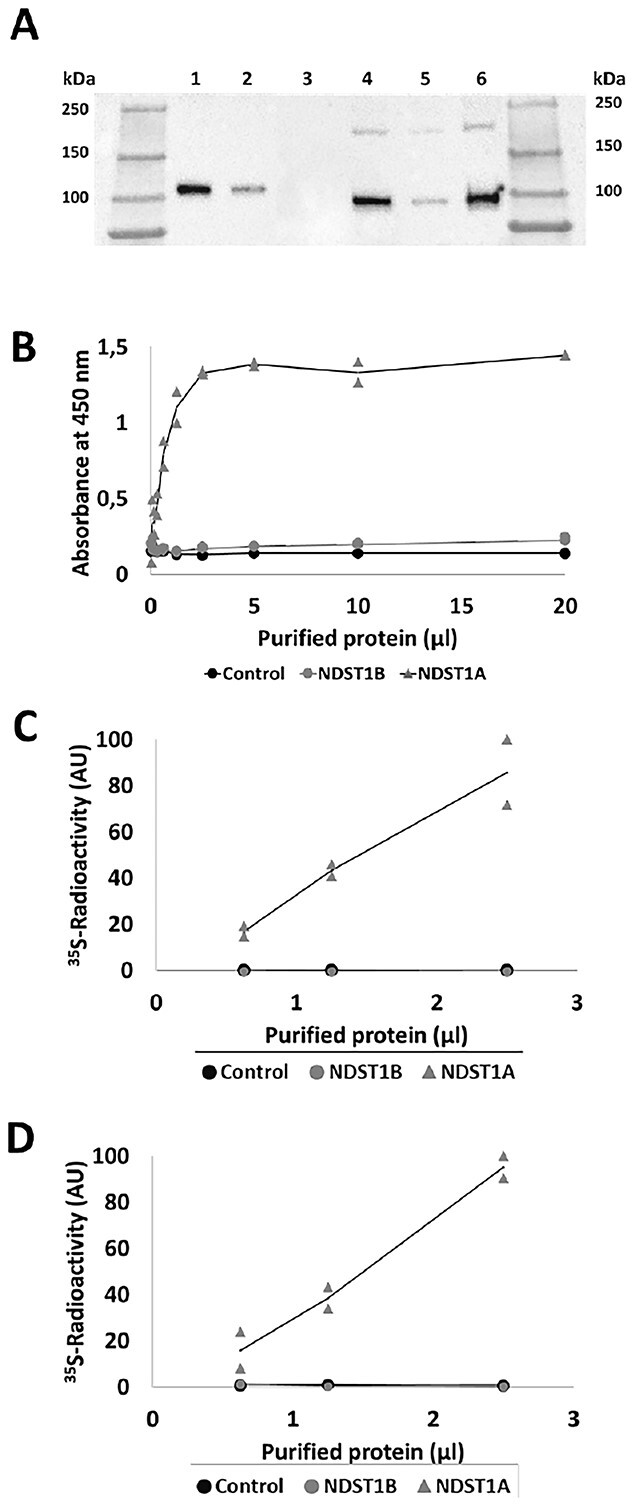
Affinity purification and enzyme activities of recombinant NDST1A and NDST1B. (A) SDS-PAGE followed by western blotting of purified NDST1 proteins detected with α-DDK (FLAG) mouse monoclonal antibody. An eluate from anti-FLAG-tag magnetic beads previously incubated with a lysate of untransfected HEK 293 cells was used as a control. Lane 1: 10 μL NDST1A; lane 2: 2.5 μL NDST1A; lane 3: 10 μL “control”; lane 4: 10 μL NDST1B; lane 5: 2.5 μL NDST1B; lane 6: 20 μL NDST1B. Note that the protein concentration in the 200 μL eluates was too low to allow for determination of protein concentration. (B) N-deacetylase ELISA assay of serial dilutions of purified protein from HEK 293 cells as control (•), and transfected with NDST1A ( ) and NDST1B (
) and NDST1B ( ), respectively. (C) N-sulfotransferase activity and (D) combined N-deacetylase and N-sulfotransferase activities. The values shown are the mean of duplicates.
), respectively. (C) N-sulfotransferase activity and (D) combined N-deacetylase and N-sulfotransferase activities. The values shown are the mean of duplicates.
NDST1B affects HS composition and domain structure
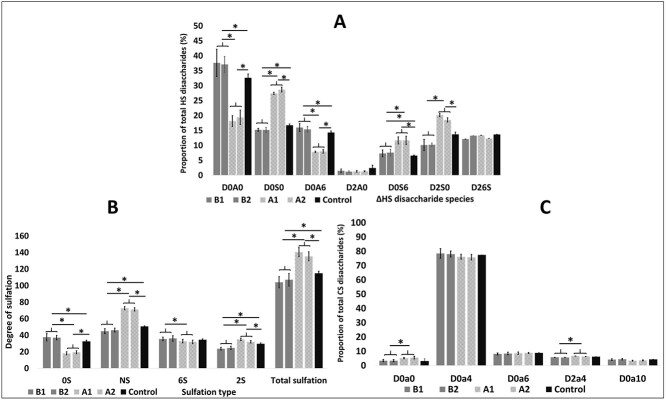
Previous studies have shown that overexpression of full length mouse NDST1 results in increased HS sulfation (Pikas et al. 2000; Presto et al. 2008). This result was confirmed for the human enzyme when HS disaccharide analysis was performed on NDST1A overexpressing HEK293 cells (Fig. 5A and B). Interestingly, the opposite was seen in HEK 293 cells overexpressing NDST1B (Fig. 5A and B), demonstrating that the lowered NDST enzyme activity in these cells affected HS biosynthesis; Total N-sulfation and 2-O-sulfation were decreased (Fig. 5A), mostly caused by a reduction in mono N-sulfated disaccharides and in disaccharides substituted with both an N- and a 2-O-sulfate group (Fig. 5B). As a compensation, the levels of 6-O-sulfated disaccharides were slightly increased (Fig. 5B). Chondroitin sulfate composition was not affected by NDST1B overexpression while a slight increase in CS 4-O-sulfation was observed in the NDST1A overexpressing cells (Fig. 5C). To confirm the ability of NDST1B to affect HS biosynthesis, the disaccharide composition of HS from HeLa cells transiently transfected with the NDST1B construct was also tested (Supplementary Fig. S1). Similar to HS isolated from HEK293 cells, a larger portion of the disaccharides from HeLa cell HS were non-sulfated. However in HeLa cell HS, also 6-O-sulfation was decreased.
Fig. 5.
HS and CS disaccharide composition. Disaccharide composition of HS (A and B) and CS (C) produced by HEK 293 cell lines overexpressing NDST1A (light gray bars) or NDST1B (dark gray bars) or transfected with empty vector (black bars) analyzed by RPIP-HPLC. The values for the different cell lines shown are the mean of triplets ± SD. An asterisk * indicates a significant difference (P < 0.05), calculated using the 2-tailed Student’s t-test. (A) HS disaccharide composition: D0A0, HexAGlcNAc; D0S0, HexAGlcNS; D0A6, HexAGlcNAc(6S); D2A0, HexA(2S) GlcNAc; D0S6, HexAGlcNS(6S); D2S0, HexA(2S)GlcNS; D26S, HexA(2S) GlcNS(6S). (B) Percentage of unsulfated disaccharides (OS), N-sulfated disaccharides (NS), 6-O-sulfated disaccharides (6S), and 2-O-sulfated disaccharides (2S). Total sulfation is the sum of N-sulfate, 2-O-sulfate, and 6-O-sulfate groups in 100 disaccharides. (C) CS disaccharide composition: D0a0: HexAGalNAc; D0a4: HexAGalNAc(4S); D0a6: HexAGalNAc(6S); D2a4: HexA(2S)GalNAc(4S); D0a10 HexAGalNAc4S6S. Disaccharide abbreviations are according to the published structural code in Lawrence et al. (2008).
To study the effects of the lowered N-sulfation in the NDST1B overexpressing HEK 293 cells on HS domain structure, HS metabolically labeled with [3H]glucosamine isolated from control cells and cells overexpressing NDST1B, respectively, was treated with HNO2 at pH 1.5 followed by separation of the cleavage products on a Bio-Gel P-10 column (Fig. 6A). [3H]Glucosamine-labeled HS from NDST1A overexpressing and HEK 293 control cells were also analyzed in the same fashion (Fig. 6B). Since HNO2 cleaves HS at every GlcNS residue, the cleavage pattern can be used to calculate overall N-sulfation but it also gives information about distribution of the N-sulfate groups (Dagalv et al. 2015). P10 gel filtration of the NDST1A HS demonstrated that disaccharides and tetrasaccharides were the main products (Fig. 6B), while the NDST1B sample contained relatively more of the tetrasaccharide peak (Fig. 6A). Interestingly, the cleavage pattern of HS in the NDST1B overexpressing cells was different from that of control HS (Fig. 6A). In comparison with the cleaved HS from control cells, the tetrasaccharide peak as well as the hexa- and octa-saccharide peaks were larger in the cleaved HS from NDST1B overexpressing cells, while the disaccharide peak was smaller, indicating that the NS-domains in this HS may be shorter than in HS from control cells (see model in Fig. 6C).
Fig. 6.
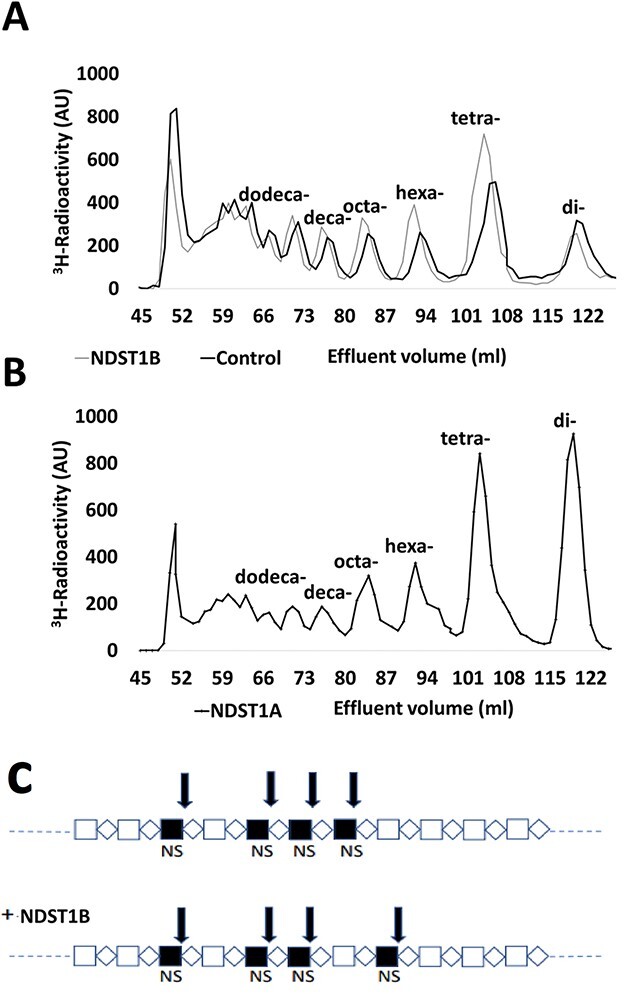
HS domain structure. Gel chromatography on bio-gel P-10 of 3H-labeled HS after treatment of the polysaccharide with nitrous acid at pH 1.5. (A) HS from control HEK 293 cells (black line) and from cells overexpressing NDST1B (gray line). (B) HS from cells overexpressing NDST1A. (C) Tentative model of an NS-domain in HEK 293 cells (top) and in HEK 293 cells overexpressing NDST1B (bottom). The arrows indicate HNO2-sensitive cleavage sites. Note that 2 disaccharides and 1 tetrasaccharide will be generated from the upper saccharide after HNO2 treatment, while the lower saccharide instead will be cleaved into 1 disaccharide and 2 tetrasaccharides. In addition, 2 oligosaccharides of larger size will be generated from both saccharides.
Enzyme–enzyme interactions
It has become increasingly clear that enzyme–enzyme interactions take place in the Golgi compartment and that they have great impact on glycan synthesis. Using coimmunoprecipitation, we have previously shown that NDST1 (NDST1A) interacts with EXT2 (Presto et al. 2008), but lack of high quality EXT1 antibodies have prevented us from investigating NDST–EXT1 interactions. Using fluorescence resonance energy transfer (FRET)-microscopy, we now show that NDST1A can bind to EXT1 and that this ability is shared by NDST1B (Fig. 7). Likewise, the ability of NDST1A to interact with EXT2 was also shared with NDST1B (Fig. 7).
Fig. 7.
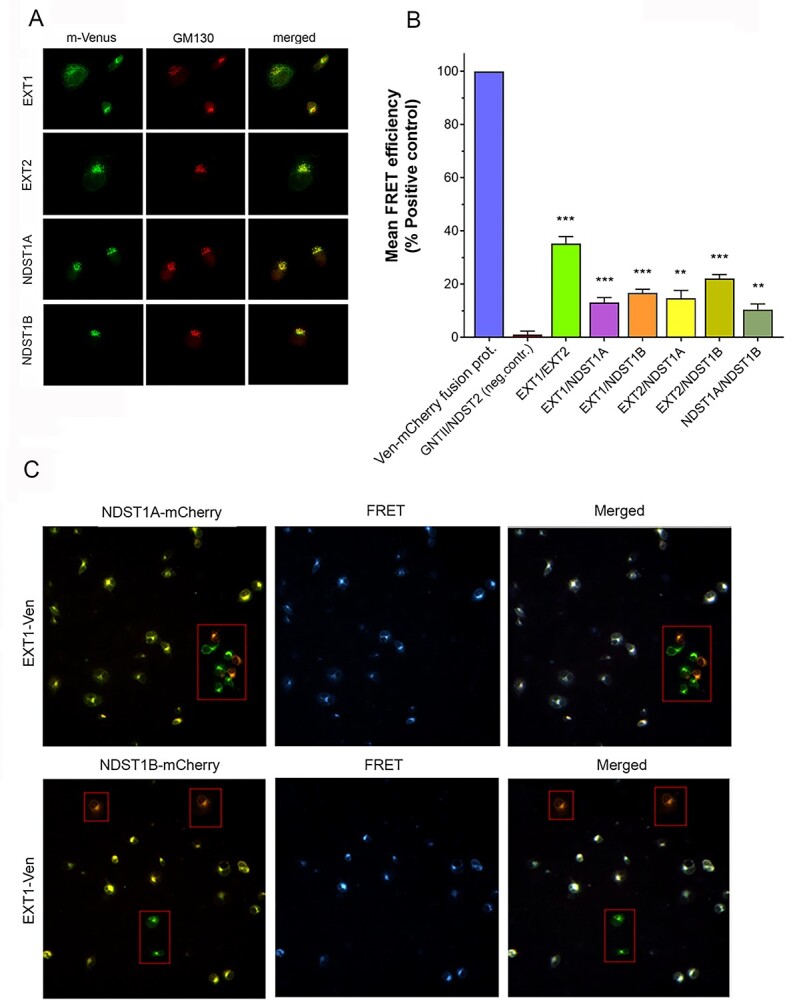
Mutual interactions of EXT1, EXT2, NDST1A, and NDST1B in the Golgi compartment in transfected COS-7 cells. (A) Coimmunostaining of the Venus-tagged EXT1, EXT2, NDST1A, and NDST1B with the cis-Golgi marker protein GM130 in COS-7 cells. For visualization of GM130, a specific antibody and Alexa 594 secondary antibody were used. (B) FRET signals measured after coexpression of the depicted mVenus and mCherry-tagged enzyme constructs. FRET efficiencies were normalized against the positive control values (blue bar, the mVenus–mCherry fusion protein), and presented as percentages of positive control. As a negative control, an O-glycosylation enzyme (GNTII) and NDST2 was used as a FRET pair. For each FRET pair, the mean FRET efficiency was determined in triplicate (± SD, n = 3) from 12,000 cells in each case (**, P < 0.01; ***, P < 0.001). (C) The localization and specificity of the FRET signal in the transfected cells. Fluorescence microscopy of the COS-7 cells transfected with EXT1-Venus/NDST1A-mCherry (top row) or EXT1/NDST1B-mCherry (bottom row) enzyme pairs. Note that cells express either both (yellow) or only one of the constructs (red, green; boxed areas) after merging the Venus and mCherry channels (left panel). FRET signal (pseudocolored as blue) from the same cells is shown in the middle panel. Superimposition of the 2 channels (right panel) shows that only cells that express both constructs display FRET signal (white dots), while those expressing only one of the constructs do not (boxed cells). The presence of cells that express only 1 of the 2 constructs constitute 10–15% of the transfected cells. The true FRET efficiency percentages are therefore somewhat higher than those shown in (B).
NDST1B and cancer
So, besides serving as a tool to understand HS biosynthesis, does NDST1B have any physiological or pathological function? Removing exon 12 is obviously a quick way to downregulate NDST activity and generate an inactive enzyme with an impact on HS biosynthesis. By exploring data from The Cancer Genome Atlas (TCGA) project (Cancer Genome Atlas Research et al. 2013), we found that high expression of NDST1B in adrenocortical carcinoma, kidney renal clear cell carcinoma, mesothelioma and uveal melanoma is associated with shorter survival (Fig. 8A). A significantly increased expression of NDST1B compared to normal tissue is also seen in several other types of cancer (Fig. 8B).
Fig. 8.
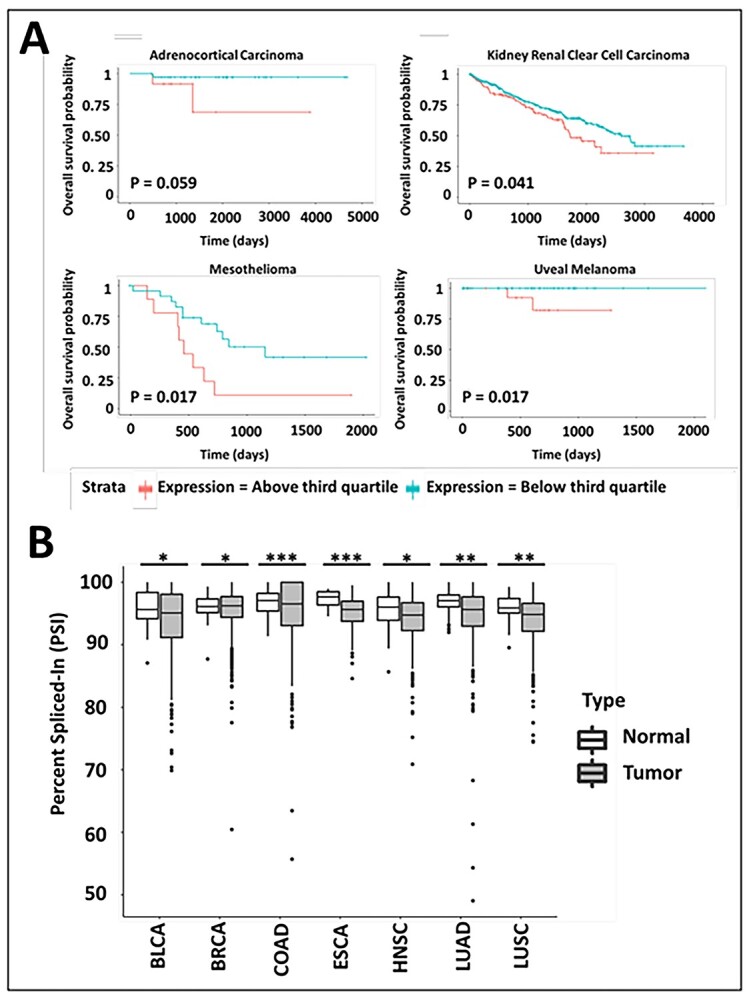
NDST1B expression in cancer. (A) NDST1B expression and overall survival probability of patients involved in TCGA project (Cancer Genome Atlas Research et al. 2013). Data were downloaded from TSVdb (Sun et al. 2018) and a log-rank test was used for calculating difference between NDST1b transcript expression groups with expression above and below the third quartile levels. (B) Data describing significant exon skipping of NDST1 exon 12 in tumors and their corresponding normal tissues from TCGA project (Cancer Genome Atlas Research et al. 2013). The percent spliced-in value indicates the number of transcripts including exon 12. Data were downloaded from TCGASpliceSeq (Ryan et al. 2016) and a 2-tailed Student’s t-test were used for calculating significant differences. Asterisk * indicates a significant difference * = P ≤ 0.05, ** = P ≤ 0.01, *** = P ≤ 0.001. BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; COAD, colon adenocarcinoma; ESCA, Esophageal carcinoma; HNSC, head and neck squamous cell carcinoma; LUAD lung adenocarcinoma; LUSC, lung squamous cell carcinoma.
Discussion
NDST1A and NDST1B are the 2 main transcripts of the human NDST1 gene. Exon 12, absent in NDST1B, encodes aa 716–772 (see Fig. 1A). This part of the protein is found in the C-terminal half of the protein which contains the N-sulfotransferase active site. The N-sulfotransferase part of the protein has been crystallized (Kakuta et al. 1999, 2003) but crystallization of the entire protein has so far failed. Interestingly, modeling of the structure of the C-terminal part of the protein with a hexasaccharide substrate indicated that His716 and His720 take part in substrate binding. In the 2003 paper (Kakuta et al. 2003), it was also shown that mutation of either of these residues to alanine resulted in drastically reduced N-sulfotransferase activity. Since both histidines are absent in NDST1B, the lack of N-sulfotransferase activity of NDST1B could be expected (Fig. 4C). However, NDST1B also lacked N-deacetylase activity (Fig. 4B) and no enzyme activity could be detected with the combined assay (Fig. 4C). Thus, this part of the protein seems to be directly or indirectly involved also in N-deacetylation. Interesting to note is that one of the mutations in the patient with NDST1 syndrome, A736V, is located in the region that is absent in NDST1B (Armstrong et al. 2017).
Unexpectedly at first, enzyme activities in lysates of cells overexpressing NDST1B were consistently lower than in mock-transfected HEK 293 cells (Figs 3 and 4). The decreased activity also affected the structure of HS produced by the cells, resulting in lowered sulfation (Fig. 5). While N-sulfation and 2-O-sulfation were decreased, a small but significant increase in 6-O-sulfation could be detected. These structural alterations could tentatively all be due to a lowered N-sulfation since 2-O-sulfation depends on epimerization of GlcA into IdoA which occurs in N-sulfated regions (Kreuger and Kjellen 2012) and 6-O-sulfation often compensates for lost N- and/or 2-O-sulfation (Merry et al. 2001; Ringvall and Kjellen 2010). In addition, the domain structure of HS was influenced by NDST1B overexpression, tentatively resulting in shorter N-sulfated domains (Fig. 6).
We have previously shown that NDST1 can form a complex with EXT2 and that overexpression of EXT2 results in increased HS N-sulfation (Presto et al. 2008). If NDST1B also has the ability to interact with EXT2, it may compete with endogenous NDST1A for binding to EXT2 and thereby inhibit N-sulfation. Using high-content FRET-microscopy analysis of cells expressing enzymes tagged with donor and acceptor fluorophores, we indeed found that NDST1B has the capacity to interact with EXT2 (Fig. 7). The result demonstrating that EXT1 also may serve as a binding partner for both NDST1A and NDST1B calls for more experiments to further understand the interplay between the enzymes.
The potential role of enzyme–enzyme interactions has long been discussed in the field and enzyme complexes have previously been shown to exist for N-glycan trimming N-acetylglycosyltransferases (36) (Khoder-Agha et al. 2019a, 2019b). In an article published in 2002, Esko and Selleck (2002) coined the term “GAGosome” to describe a tentative enzyme machinery responsible for elongation and modification of glycosaminoglycans. Even though the presence of a big complex comprising all enzymes seems less likely, several investigations have reported on interactions between 2 (Schwartz et al. 1974; Kobayashi et al. 2000; McCormick et al. 2000; Pinhal et al. 2001), and more rarely, 3 enzymes (Dejima et al. 2013). For instance, it was demonstrated that the polymerases EXT1 and EXT2 form a functional complex and are more active when they are co-expressed (Senay et al. 2000). Interaction between HS C-5 epimerase and the HS 2-O-sulfotransferase (Pinhal et al. 2001) are other examples.
Our data demonstrates that the enzymatically dead NDST1B splice variant still has the capacity to influence HS structure, most likely by competing with the endogenous active NDST1 and replacing it in the enzyme complex. These results are important when it comes to understanding the regulation of HS biosynthesis in general; HS structure is influenced by the composition of the enzyme complexes; Exchanging one enzyme for another will alter the structure according to the loss or gain of the enzymatic activities but also indirectly through the substrate specificities of the enzymes acting after the exchanged enzyme. So, when an active NDST is replaced by an inactive, N-sulfation will decrease, but also epimerization of GlcA into IdoA which is the preferred substrate for 2-O-sulfation. Why expression of NDST1B in HEK293 cells resulted in slightly increased 6-O-sulfation while a decrease instead was seen in HeLa cells could tentatively be due to different expression of the 3 6-O-sulfotransferases in the 2 cell types. Another illustration of the effect of substituting an enzyme for another is the increased sulfation of mast cell heparin as a result of lowered expression of NDST1 (Dagalv et al. 2011); The tentative explanation of this finding is that the more efficient NDST2 in the absence of NDST1 could occupy more of the actively synthesizing enzyme complexes in the cell.
It is interesting that high expression of NDST1B is associated with shorter survival of patients with different types of cancer and that a significantly increased expression of NDST1B compared to normal tissue is seen in several other types of cancer (Fig. 8). Even though the lowering effect on HS sulfation is modest, it will result in altered binding of growth factors/cytokines (Kjellen and Lindahl 2018), which could alter the growth and metastatic properties of the cells. Future studies will hopefully reveal to what degree alternative splicing of HS biosynthesis enzymes will contribute to cancer pathology.
Materials and methods
cDNA constructs
For transfection of HEK 293 and HeLa cells
A pCMV6-vector containing cDNA (RC221638) encoding human NDST1(NM__01543) with a C-terminal Myc/Flag-tag was obtained from Origene. The corresponding protein product is in this report denoted NDST1A. This vector was also used to generate the shorter form of NDST1, here denoted NDST1B, through a Quick Change mutagenesis approach removing the cDNA corresponding to exon 12 (see Fig. 1). The primers were designed to delete 171 bp at the position marked with an asterisk:
NDST1B QC fw: CTATTCCTGGTACCAG*ATTCTGGTCTTGGA
NDST1B QC rv: TCCAAGACCAGAAT*CTGTTACCAGGAATAG
The mutant version was amplified using Phusion High FidelityPolymerase (Thermo) according to the manufacturer’s instructions (30 s at 98 °C, 20 cycles of 10 s at 98 °C and 3:45 min at 72 °C, finally 10 min at 72 °C). Following DNA amplification the remaining template was digested with DpnI (Thermo) and the PCR product was transformed into TOP10 cells (Thermo) following the manufacturer’s instructions. Introduction of the desired mutation was confirmed by sequencing before the construct was used for transfection of cells.
For FRET interaction assays
Plasmids for enzyme interaction study (FRET) were all based on a pcDNA3 vector backbone (Invitrogen) and the constructs were prepared using the full-length cDNA clones for NDST1A and NDST1B described above, a full-length clone for human NDST2 with a C-terminal Myc/Flag-tag (from Origene) and plasmids expressing full-length human C-terminal myc-DDK-tagged EXT1 in the pCMV6-entry vector and Turbo green fluorescent protein (GFP) (tGFP)-tagged EXT2 in the pCMV6-AC-GFP vector (both from Origene) using PCR amplification with specific primers (see below). For each enzyme, the PCR product was inserted into the pcDNA3 vector and the sequence was verified before use. The vector also contained sequences encoding a C-terminal monomeric Venus (mVen) and monomeric Cherry (mChe) (Khoder-Agha et al. 2019b).
EXT1 EcoRI fw: ATTGCGAATTCATGCAGGCCAAAAAACGC
EXT1 XbaI rv: ATTGCTCTAGAAAGTCGCTCAATGTCTCGG
EXT2 EcoRI fw: ATTGCGAATTCATGTGTGCGTCGGTCAAG
EXT2 XbaI rv: ATTGCTCTAGATAAGCTGCCAATGTTGGGG
NDST1A/B EcoRI fw: ATTGCGAATTCATGCCTGCCCTGGCATG
NDST1A/B XbaI rv: ATTGCTCTAGACCTGGTGTTCTGGAGGTCCTC
NDST2 EcoRI fw: ATTGCGAATTCATGCTCCAGTTGTGGAAGG
NDST2 XbaI rv: ATTGCTCTAGAGCCCAGACTGGAATGCTG
Transfection of HEK 293 and HeLa cells
Cells (human embryonic kidney (HEK) 293 cells or HeLa cells) were grown in Dulbecco’s Modified Eagle Medium (Gibco DMEM) containing 10% of fetal bovine serum and 60 μg/mL penicillin and 50 μg/mL streptomycin. HEK 293 cell lines overexpressing NDST1A and NDST1B, were generated by transfection of the pCMV6-vectors using Lipofectamine2000 reagent from Invitrogen according to the manufacturer’s protocol. GFP plasmid was used as a positive control for monitoring the transfection efficiency. The pCMV6 vector carries neomycin resistance and stable clones were selected using Geneticin (G418) (Gibco). A high level of G418, 1 mg/mL, was used for selection of the clones while selected clones were maintained at a concentration of 0.2 mg/mL G418. Transient transfection of HeLa cells was carried out using Lipofectamine3000 reagent from Invitrogen according to the manufacturer’s protocol.
Solubilization of HEK 293 cells
Cells were harvested at 90% confluency. After washing twice with PBS, the cells were gently washed off the T75 flasks in PBS and transferred to a 15 mL tube followed by centrifugation at 1000 rpm for 4 min to pellet the cells. The pelleted cells were stored at −20 °C. Cell lysates were generated by addition of cell lysis buffer (50 mM Tris–HCl, pH 7.4, 0.15 M NaCl, 1% Triton X-100 and EDTA-free protease inhibitor cocktail (Roche) to the pellets (1 mL/cells recovered from a T75 flask) followed by end-over-end incubation for 1 h at 4 °C. After centrifugation at 13,200 rpm, at 4 °C for 15 min, the supernatants were recovered and assayed directly for enzyme activities or used for purification of the recombinant proteins. Aliquots were also used for determination of protein concentration and for SDS-PAGE followed by western blotting.
Purification of recombinant proteins
The lysates were passed through Filtropur Syringe Filters, 0.45 μm pore size (Sarstedt) before addition to 100 μL slurry/mL lysate of anti-FLAG-tag magnetic beads (Sigma-Aldrich), previously washed with TBS. After end-over-end incubation for 1 h at 4 °C, the “unbound” fraction was recovered and the beads were washed at least 3 times with 1 mL 50 mM Tris–HCl pH 8, 138 mM NaCl and 2.7 mM KCl, 1% Triton X-100 and 1% Roche protease inhibitor cocktail. After the last washing step, the absorbance was measured to check for remaining protein. When the absorbance at 280 nm of the last washing was <0.05 the beads were eluted with 200 μL portions of 2% Flag-peptide (Sigma-Aldrich) dissolved in the washing buffer.
SDS-PAGE and western blotting
Cell lysates corresponding to 40 μg of protein (Fig. 1) and aliquots from fractions obtained during the purification procedure (Fig. 4) were run on 4–15% mini-PROTEAN SDS-polyacrylamide gels (Bio-Rad) followed by blotting to nitrocellulose membranes. Next the membranes were blocked with 5% milk powder in TBS-T for 60 min and then incubated with the indicated primary antibodies (rabbit anti-trunc-NDST1; Presto et al. 2008), diluted 1:1,000 in blocking buffer or anti-DDK (FLAG) mouse monoclonal antibody (ORIGENE), diluted 1:2,000 in blocking buffer). After the membranes had been washed in TBS-T they were incubated in blocking buffer with secondary antibody (goat anti-rabbit horseradish peroxidase antibodies, diluted 1:5,000 and goat anti-mouse horseradish peroxidase antibodies, diluted 1:4,000, respectively) for 60 min at room temperature. The membranes were then extensively washed and the peroxidase activity was detected using the Prime-ECL reagent (GE Healthcare) captured with a Bio-Rad Western Blot Imaging System.
NDST enzyme activity assays
Oligosaccharides derived from Escherichia coli K5 capsular polysaccharide, composed of alternating GlcA-GlcNAc-residues, were used as substrate in the N-deacetylase and the combined N-deacetylase/N-sulfotransferase assays. N-Sulfotransferase activity was analyzed using an N-deacetylated K5 polysaccharide as substrate. All samples from at least 2 independent experiments were analyzed in duplicate with the 3 different assays described below.
N-Deacetylase ELISA assay: This assay was carried out as previously described (Dagalv et al. 2015). Briefly, a 96 flat bottom well (Nunc MaxiSorp) plate was coated with K5 polysaccharide. After washing with TBS-T, cell lysates were added and the plate was incubated at 37 °C for 30 min followed by washing with TBS-T. After blocking with 1% gelatin in TBS, the JM 403 antibody recognizing the deacetylated substrate (van den Born et al. 2003) was added to the wells followed by incubation for 1 h at room temperature. After washing, the wells were incubated with goat anti-mouse IgM-peroxidase solution for 1 h at room temperature. Peroxidase substrate mix was then added after washing. The reaction was stopped after 10–15 min with 2 M H2SO4 and absorbance at 450 nm was measured in a spectrophotometer plate reader.
N-Sulfotransferase and combined N-Deacetylase/N-Sulfotransferase assay: N-Sulfotransferase activity was analyzed by measuring incorporation of 35S from the sulfate donor [35S]PAPS into N-deacetylated K5 polysaccharide substrate. Cell lysates or purified recombinant proteins were incubated with substrate and 2 μCi of [35S]PAPS in 50 mM Hepes, pH 7.4, 10 mM MgCl2, 10 mM MnCl2, 5 mM CaCl2, 3.5 μM NaF, and 1% Triton X-100 in 0.1 mL for 30 min at 37 °C. The polysaccharide substrate was precipitated with ethanol for 4 h, separated from excess [35S]PAPS by Sephadex G-25 superfine (GE Healthcare) gel filtration, and quantified by scintillation counting (Dagalv et al. 2015). The same protocol was followed for the combined N-deacetylase/N-sulfotransferase assay, but the substrate was exchanged for K5 polysaccharide which had not been N-deacetylated.
Metabolic labeling and isolation of 3H-labeled heparan sulfate
The HEK 293 cell lines were incubated with 50 μCi/mL 3H-glucosamine in the cell culture medium described above at 37 °C for 24 h. 3H-labeled proteoglycans were then purified from the cell lysates after DEAE-ion exchange chromatography and gel chromatography on Superose 6 columns as previously described in detail (Dagalv et al. 2015). After alkali treatment, the released glycosaminoglycan chains were incubated with chondroitinase ABC and subjected to another gel chromatography on Superose 6 to remove 3H-labeled CS degradation products (Dagalv et al. 2015). After desalting on PD-10 columns (GE Healthcare) the 3H-labeled HS-chains were ready for analysis of domain structure.
Analysis of HS domain structure
3H-Labeled purified HS chains (20,000 cpm) were treated with nitrous acid at pH 1.5, which cleaves the polysaccharide at GlcNS residues (Shively and Conrad 1976), followed by size-dependent separation of the generated fragments on a 1 × 200 cm Bio-Rad Bio-gel P-10 Fine column eluted with 0.2 M NH4HCO3. Heparin was used as a carrier and dextran blue and [3H] H2O were used as markers for the void volume and total volume, respectively. 155 fractions of 1 mL were collected and analyzed for radioactivity (Dagalv et al. 2015).
GAG compositional analysis
Glycosaminoglycans were isolated from the HEK 293 cell lines or the transiently transfected HeLa cells as described previously (Ledin et al. 2004 and modified by Dagalv et al. 2011). Chondroitin sulfate and HS disaccharides generated after digestion with chondroitinase ABC and heparinase I, II, and III, respectively, were subjected to RPIP-HPLC analysis followed by postcolumn derivatization with cyanoacetamide and detection in a fluorescence detector (Ledin et al. 2004).
High-content FRET interaction assay and quantification
COS-7 cells were transfected with Lipofectamine 3000 kit (Invitrogen) with the relevant FRET plasmid. Cells were seeded onto Cell Carrier Ultra 96-well plates and allowed to attach for 6 h. 400 ng/ mL of each construct, mVenus (mVen, donor), and mCherry (mChe, acceptor), was used for transfection of 10,000 cells per well. 18 h after post transfection, cells were fixed with 4% paraformaldehyde (PFA) and washed with PBS (pH 7.4). Using Operetta (Perkin-Elmer Inc.), with appropriate filter sets for mVenus and mCherry, FRET signals were measured. Quantification was performed with Harmony 4.6 software using the Youvan FRET formula (Hassinen et al. 2011). Data are shown as the average FRET signal from 2,000 cells/well, performed in 6 wells each in triplicates (± S.D., no. of wells = 18).
Colocalization studies with fluorescence microscopy
Colocalization studies were performed in the cells expressing the selected mVenus-tagged enzyme constructs as described previously (Kokkonen et al. 2004). 20 h post-transfection, cell were fixed as above and stained with the antibody against the endogenous cis-Golgi marker protein GM130 and species-specific Alexa fluor-594-conjugated anti-mouse and anti-rabbit secondary antibodies (Invitrogen, Carlsbad, CA, USA), mounted and imaged using the Olympus BX51 microscope (Olympus Optical Co., Ltd, Japan) equipped with a 60X Plan-Apo oil-immersion objective and appropriate filter sets for each dye.
Supplementary Material
Contributor Information
Parisa Missaghian, Department of Medical Biochemistry and Microbiology, The Biomedical Center, Box 582, SE-75123 Uppsala, Sweden.
Tabea Dierker, Department of Medical Biochemistry and Microbiology, The Biomedical Center, Box 582, SE-75123 Uppsala, Sweden.
Elham Khosrowabadi, Faculty of Biochemistry and Molecular Medicine, Aapistie 7A, 90220 Oulu, Finland.
Fredrik Axling, Department of Surgical Sciences, Uppsala University Hospital, SE-751 85 Uppsala, Sweden.
Inger Eriksson, Department of Medical Biochemistry and Microbiology, The Biomedical Center, Box 582, SE-75123 Uppsala, Sweden.
Abdurrahman Ghanem, Department of Medical Biochemistry and Microbiology, The Biomedical Center, Box 582, SE-75123 Uppsala, Sweden.
Marion Kusche-Gullberg, Department of Biomedicine, University of Bergen, N-5009 Bergen, Norway.
Sakari Kellokumpu, Faculty of Biochemistry and Molecular Medicine, Aapistie 7A, 90220 Oulu, Finland.
Lena Kjellén, Department of Medical Biochemistry and Microbiology, The Biomedical Center, Box 582, SE-75123 Uppsala, Sweden.
Funding
This work was supported by The Swedish Cancer Society Grant number 20 1338 PjF, Stiftelsen för Proteoglykanforskning, and the Medical Faculty at Uppsala University.
Conflict of interest statement
None declared.
Abbreviations
FRET, fluorescence resonance energy transfer; GlcA, glucuronic acid; GlcNAc, N-acetylglucosamine; GlcNH2, glucosamine; GlcNS, N-sulfoglucosamine; HS, heparan sulfate; NDST, glucosaminyl N-deacetylase/N-sulfotransferase; PAPS, 3′-phosphoadenosine 5′-phosphosulfate; TCGA, The Cancer Genome Atlas
References
- Ai X, Do AT, Kusche-Gullberg M, Lindahl U, Lu K, Emerson CP Jr. 2006. Substrate specificity and domain functions of extracellular heparan sulfate 6-O-endosulfatases, QSulf1 and QSulf2. J Biol Chem. 281:4969–4976. [DOI] [PubMed] [Google Scholar]
- Aikawa J, Grobe K, Tsujimoto M, Esko JD. 2001. Multiple isozymes of heparan sulfate/heparin GlcNAc N-deacetylase/GlcN N-sulfotransferase. Structure and activity of the fourth member, NDST4. J Biol Chem. 276:5876–5882. [DOI] [PubMed] [Google Scholar]
- Armstrong L, Tarailo-Graovac M, Sinclair G, Seath KI, Wasserman WW, Ross CJ, van Karnebeek CD. 2017. A girl with developmental delay, ataxia, cranial nerve palsies, severe respiratory problems in infancy-expanding NDST1 syndrome. Am J Med Genet A. 173:712–715. [DOI] [PubMed] [Google Scholar]
- van den Born J, Pikas DS, Pisa BJ, Eriksson I, Kjellen L, Berden JH. 2003. Antibody-based assay for N-deacetylase activity of heparan sulfate/heparin N-deacetylase/N-sulfotransferase (NDST): novel characteristics of NDST-1 and -2. Glycobiology. 13:1–10. [DOI] [PubMed] [Google Scholar]
- Bulow HE, Hobert O. 2006. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 22:375–407. [DOI] [PubMed] [Google Scholar]
- Busse-Wicher M, Wicher KB, Kusche-Gullberg M. 2014. The exostosin family: proteins with many functions. Matrix Biol. 35:25–33. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N, Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. 2013. The cancer genome atlas pan-cancer analysis project. Nat Genet. 45:1113–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagalv A, Holmborn K, Kjellen L, Abrink M. 2011. Lowered expression of heparan sulfate/heparin biosynthesis enzyme N-deacetylase/n-sulfotransferase 1 results in increased sulfation of mast cell heparin. J Biol Chem. 286:44433–44440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagalv A, Lundequist A, Filipek-Gorniok B, Dierker T, Eriksson I, Kjellen L. 2015. Heparan sulfate structure: methods to study N-sulfation and NDST action. Methods Mol Biol. 1229:189–200. [DOI] [PubMed] [Google Scholar]
- Dejima K, Takemura M, Nakato E, Peterson J, Hayashi Y, Kinoshita-Toyoda A, Toyoda H, Nakato H. 2013. Analysis of drosophila glucuronyl C5-epimerase: implications for developmental roles of heparan sulfate sulfation compensation and 2-O-sulfated glucuronic acid. J Biol Chem. 288:34384–34393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deligny A, Dierker T, Dagalv A, Lundequist A, Eriksson I, Nairn AV, Moremen KW, Merry CLR, Kjellen L. 2016. NDST2 (N-deacetylase/N-Sulfotransferase-2) enzyme regulates heparan sulfate chain length. J Biol Chem. 291:18600–18607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delos M, Foulquier F, Hellec C, Vicogne D, Fifre A, Carpentier M, Papy-Garcia D, Allain F, Denys A. 2018. Heparan sulfate 3-O-sulfotransferase 2 (HS3ST2) displays an unexpected subcellular localization in the plasma membrane. Biochim Biophys Acta Gen Subj. 1862:1644–1655. [DOI] [PubMed] [Google Scholar]
- Esko JD, Selleck SB. 2002. Order out of chaos: assembly of ligand binding sites in heparan sulfate. Annu Rev Biochem. 71:435–471. [DOI] [PubMed] [Google Scholar]
- Grobe K, Ledin J, Ringvall M, Holmborn K, Forsberg E, Esko JD, Kjellen L. 2002. Heparan sulfate and development: differential roles of the N-acetylglucosamine N-deacetylase/N-sulfotransferase isozymes. Biochim Biophys Acta. 1573:209–215. [DOI] [PubMed] [Google Scholar]
- Hassinen A, Pujol FM, Kokkonen N, Pieters C, Kihlstrom M, Korhonen K, Kellokumpu S. 2011. Functional organization of Golgi N- and O-glycosylation pathways involves pH-dependent complex formation that is impaired in cancer cells. J Biol Chem. 286:38329–38340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozzo RV, Schaefer L. 2015. Proteoglycan form and function: a comprehensive nomenclature of proteoglycans. Matrix Biol. 42:11–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao TM, Li YL, Lin SW, Tzeng ST, Yu IS, Yen SJ, Tsai MH, Yang YC. 2016. Alteration of colonic epithelial cell differentiation in mice deficient for glucosaminyl N-deacetylase/N-sulfotransferase 4. Oncotarget. 7:84938–84950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakuta Y, Sueyoshi T, Negishi M, Pedersen LC. 1999. Crystal structure of the sulfotransferase domain of human heparan sulfate N-deacetylase/ N-sulfotransferase 1. J Biol Chem. 274:10673–10676. [DOI] [PubMed] [Google Scholar]
- Kakuta Y, Li L, Pedersen LC, Pedersen LG, Negishi M. 2003. Heparan sulphate N-sulphotransferase activity: reaction mechanism and substrate recognition. Biochem Soc Trans. 31:331–334. [DOI] [PubMed] [Google Scholar]
- Khoder-Agha F, Harrus D, Brysbaert G, Lensink MF, Harduin-Lepers A, Glumoff T, Kellokumpu S. 2019a. Assembly of B4GALT1/ST6GAL1 heteromers in the golgi membranes involves lateral interactions via highly charged surface domains. J Biol Chem. 294:14383–14393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoder-Agha F, Sosicka P, Escriva Conde M, Hassinen A, Glumoff T, Olczak M, Kellokumpu S. 2019b. N-acetylglucosaminyltransferases and nucleotide sugar transporters form multi-enzyme-multi-transporter assemblies in golgi membranes in vivo. Cell Mol Life Sci. 76:1821–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjellen L, Lindahl U. 2018. Specificity of glycosaminoglycan-protein interactions. Curr Opin Struct Biol. 50:101–108. [DOI] [PubMed] [Google Scholar]
- Kobayashi S, Morimoto K, Shimizu T, Takahashi M, Kurosawa H, Shirasawa T. 2000. Association of EXT1 and EXT2, hereditary multiple exostoses gene products, in golgi apparatus. Biochem Biophys Res Commun. 268:860–867. [DOI] [PubMed] [Google Scholar]
- Kokkonen N, Rivinoja A, Kauppila A, Suokas M, Kellokumpu I, Kellokumpu S. 2004. Defective acidification of intracellular organelles results in aberrant secretion of cathepsin D in cancer cells. J Biol Chem. 279:39982–39988. [DOI] [PubMed] [Google Scholar]
- Kreuger J, Kjellen L. 2012. Heparan sulfate biosynthesis: regulation and variability. J Histochem Cytochem. 60:898–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence R, Lu H, Rosenberg RD, Esko JD, Zhang L. 2008. Disaccharide structure code for the easy representation of constituent oligosaccharides from glycosaminoglycans. Nat Methods. 5:291–292. [DOI] [PubMed] [Google Scholar]
- Ledin J, Staatz W, Li JP, Gotte M, Selleck S, Kjellen L, Spillmann D. 2004. Heparan sulfate structure in mice with genetically modified heparan sulfate production. J Biol Chem. 279:42732–42741. [DOI] [PubMed] [Google Scholar]
- Lencz T, Guha S, Liu C, Rosenfeld J, Mukherjee S, DeRosse P, John M, Cheng L, Zhang C, Badner JA, et al. 2013. Genome-wide association study implicates NDST3 in schizophrenia and bipolar disorder. Nat Commun. 4:2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Wei G, Shi Z, Dryer L, Esko JD, Wells DE, Matzuk MM. 2000. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 224:299–311. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Kjellen L. 2013. Pathophysiology of heparan sulphate: many diseases, few drugs. J Intern Med. 273:555–571. [DOI] [PubMed] [Google Scholar]
- Lindahl U, Couchman J, Kimata K, Esko JD. 2015. Proteoglycans and sulfated glycosaminoglycans. In: Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Darvill AG, Kinoshita T, Packer NH, et al., editors. Essentials of glycobiology. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. p. 207–221. [Google Scholar]
- Macchi M, Magalon K, Zimmer C, Peeva E, el Waly B, Brousse B, Jaekel S, Grobe K, Kiefer F, Williams A, et al. 2020. Mature oligodendrocytes bordering lesions limit demyelination and favor myelin repair via heparan sulfate production. elife. 9:1–26. 10.7554/eLife.51735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick C, Duncan G, Goutsos KT, Tufaro F. 2000. The putative tumor suppressors EXT1 and EXT2 form a stable complex that accumulates in the Golgi apparatus and catalyzes the synthesis of heparan sulfate. Proc Natl Acad Sci U S A. 97:668–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry CL, Bullock SL, Swan DC, Backen AC, Lyon M, Beddington RS, Wilson VA, Gallagher JT. 2001. The molecular phenotype of heparan sulfate in the Hs2st−/− mutant mouse. J Biol Chem. 276:35429–35434. [DOI] [PubMed] [Google Scholar]
- Pikas DS, Eriksson I, Kjellen L. 2000. Overexpression of different isoforms of glucosaminyl N-deacetylase/N-sulfotransferase results in distinct heparan sulfate N-sulfation patterns. Biochemistry. 39:4552–4558. [DOI] [PubMed] [Google Scholar]
- Pinhal MA, Smith B, Olson S, Aikawa J, Kimata K, Esko JD. 2001. Enzyme interactions in heparan sulfate biosynthesis: uronosyl 5-epimerase and 2-O-sulfotransferase interact in vivo. Proc Natl Acad Sci U S A. 98:12984–12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presto J, Thuveson M, Carlsson P, Busse M, Wilen M, Eriksson I, Kusche-Gullberg M, Kjellen L. 2008. Heparan sulfate biosynthesis enzymes EXT1 and EXT2 affect NDST1 expression and heparan sulfate sulfation. Proc Natl Acad Sci U S A. 105:4751–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter MS, Musante L, Hu H, Diederich S, Sticht H, Ekici AB, Uebe S, Wienker TF, Bartsch O, Zechner U, et al. 2014. NDST1 missense mutations in autosomal recessive intellectual disability. Am J Med Genet A. 164A:2753–2763. [DOI] [PubMed] [Google Scholar]
- Ringvall M, Kjellen L. 2010. Mice deficient in heparan sulfate N-deacetylase/N-sulfotransferase 1. Prog Mol Biol Transl Sci. 93:35–58. [DOI] [PubMed] [Google Scholar]
- Ryan M, Wong WC, Brown R, Akbani R, Su X, Broom B, Melott J, Weinstein J. 2016. TCGASpliceSeq a compendium of alternative mRNA splicing in cancer. Nucleic Acids Res. 44:D1018–D1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarrazin S, Lamanna WC, Esko JD. 2011. Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol. 3:a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz NB, Roden L, Dorfman A. 1974. Biosynthesis of chondroitin sulfate: interaction between xylosyltransferase and galactosyltransferase. Biochem Biophys Res Commun. 56:717–724. [DOI] [PubMed] [Google Scholar]
- Senay C, Lind T, Muguruma K, Tone Y, Kitagawa H, Sugahara K, Lidholt K, Lindahl U, Kusche-Gullberg M. 2000. The EXT1/EXT2 tumor suppressors: catalytic activities and role in heparan sulfate biosynthesis. EMBO Rep. 1:282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shively JE, Conrad HE. 1976. Formation of anhydrosugars in the chemical depolymerization of heparin. Biochemistry. 15:3932–3942. [DOI] [PubMed] [Google Scholar]
- Sun W, Duan T, Ye P, Chen K, Zhang G, Lai M, Zhang H. 2018. TSVdb: a web-tool for TCGA splicing variants analysis. BMC Genomics. 19:405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.