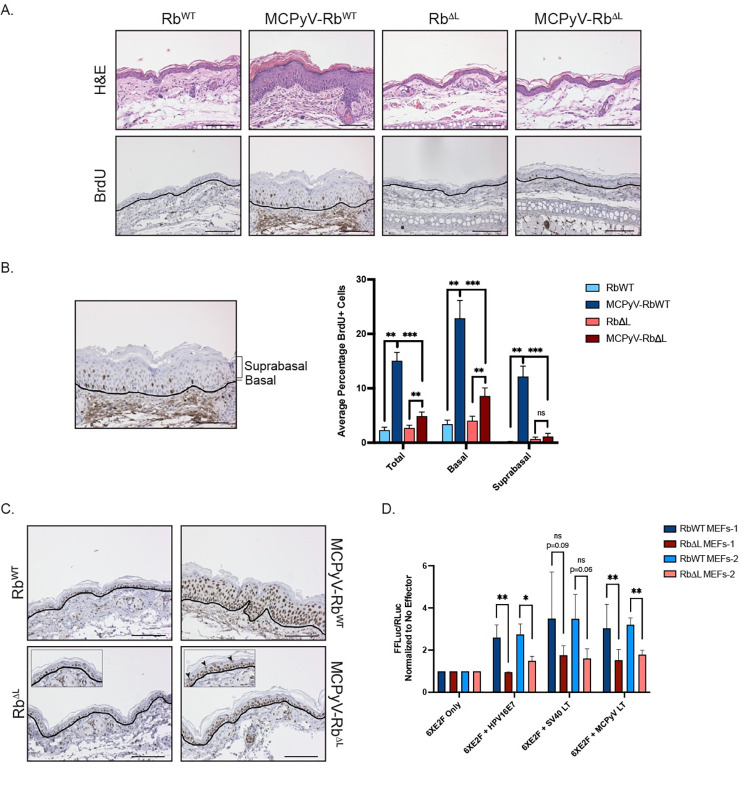
Fig 3. MCPyV T antigen-induced cellular proliferation and E2F-dependent gene expression are significantly reduced, but not eliminated, in the skin of MCPyV-RbΔL mice.
A) Ear tissues harvested from RbWT, MCPyV-RbWT RbΔL, and MCPyV-RbΔL mice were analyzed using immunohistochemistry for BrdU and counterstained with hematoxylin (bottom). BrdU-positive cells contain brown nuclei. The black line indicates the basement membrane between the epithelium and the underlying dermis. Corresponding representative H&E-stained images are included as a reference (top). All scale bars = 100 μm. B) For BrdU quantitation, basal cells were defined as any cell in contact with the basement membrane. Suprabasal cells included all other cells within the stratified epithelium that were not touching the basement membrane. An image is shown with brackets identifying the suprabasal layer and a line pointing to the basal layer. The number of total, basal, and suprabasal BrdU-positive cells in the stratified epithelium were counted in 10 fields of view per slide from each mouse per group. These values were then divided by the total number of epithelial cells in each sample to calculate the percentage of BrdU-positive cells, and these values were then averaged. An average of the averages was then calculated for each group of mice. Error bars indicate standard deviation. A Wilcoxon Rank Sum test was used to compare values among groups. Asterisks indicate statistical significance (**p≤0.002, ***p≤0.0007) and “ns” means non-significant. Please refer to the text for more information regarding statistical comparisons. C) Ear tissues harvested from RbWT, MCPyV-RbWT, RbΔL, and MCPyV-RbΔL mice were analyzed using immunohistochemistry for the MCM7 protein and counterstained with hematoxylin (bottom). MCM7-positive cells contain brown nuclei. The black line indicates the basement membrane between the epithelium and the underlying dermis. Insets are enlarged images to highlight suprabasal cells staining positive for MCM7 (black arrowheads). All scale bars = 100 μm. D) E2F-luciferase assay results from primary mouse embryonic fibroblasts (MEFs) isolated from pRbWT and pRbΔL mice transfected with viral protein effectors. MEFs from litter-matched pRbWT and pRbΔL embryos from two different litters were used: RbWT MEFs-1 and RbΔL MEFs-1 (dark blue and maroon, respectively) and RbWT MEFs-2 and RbΔL MEFs-2 (light blue and pink, respectively). Cells were transfected with an E2F-luciferase reporter construct (6XE2F; Firefly luciferase) only, or co-transfected with 6XE2F reporter plus SV40 large T antigen (LT), the MCPyV LT, or human papillomavirus 16 (HPV16) E7 protein. In all conditions, cells were co-transfected with a control plasmid expressing Renilla luciferase that was used to normalize for transfection efficiency. At 48 hours post-transfection, a dual-luciferase assay was performed to quantify firefly and Renilla luciferase expression in each condition. Following normalization of all luciferase values (firefly/Renilla), the values in cells only transfected with the reporter (6XE2F Only) were set to 1.0 and all other values were compared to this value within each MEFs cell strain. Error bars indicate standard deviation. Unpaired t-tests were used to compare luciferase values between RbWT and RbΔL cells for each condition. Asterisks indicate statistical significance (*p≤0.05, **p≤0.01) and “ns” means non-significant. Please refer to the text for more information regarding statistical comparisons.