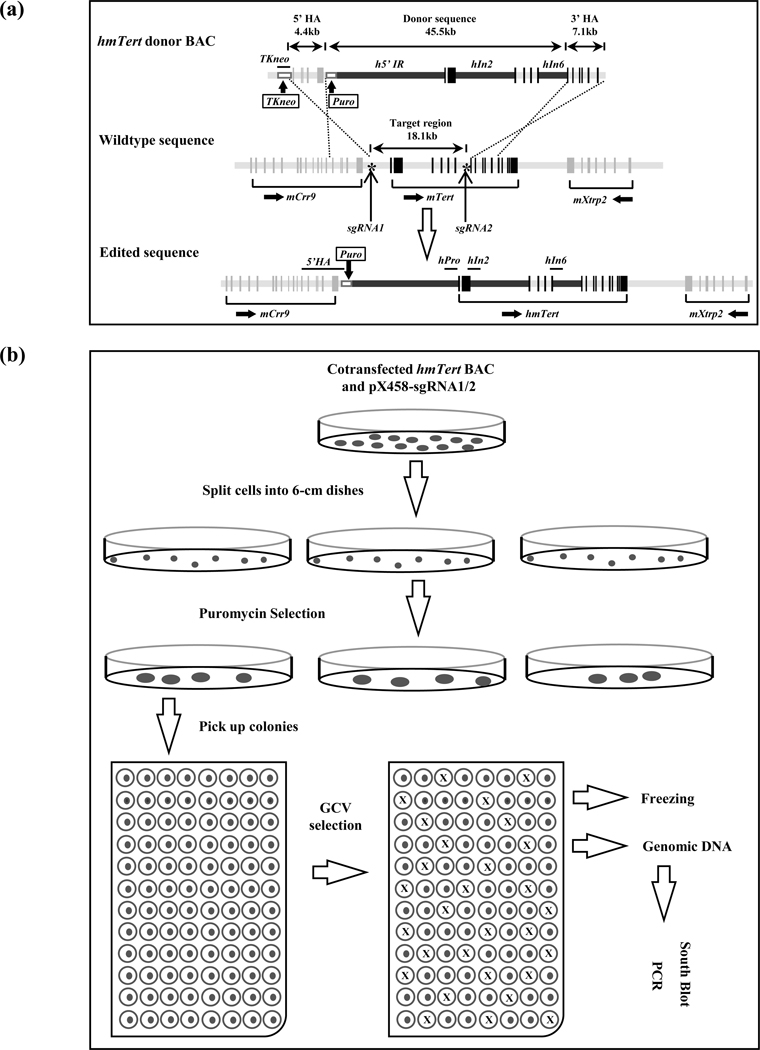
FIGURE 1.
Engineering of an hmTert allele via homologous recombination (HR). (a) Schematic illustration of the donor BAC construct and HR strategy. Sequences within the donor BAC contained the mTert gene components (light grey bars), in which its 5’IR, In2 and In6 were replaced by their counterparts from the hTERT gene (black bars). This 45.5-kb donor sequence was flanked by 5’ and 3’ homologous arms (HAs). The 5’ HA is a 4.4-kb sequence from the 3’ end of mCrr9 gene whereas the 3’ HA is a 7.1-kb region including exons 7–12 of the mTert gene. In addition, a TKneo and a puromycin-resistant cassette (white boxes) were inserted at upstream and downstream of 5’ HA, respectively. The target region is the 18.1-kb region from 5’IR to intron 6 of the mTert gene. Asterisk (*) represents Crispr/Cas9-sgRNAs targeting sites. Black vertical bars represent mTert exons. Horizontal lines above the edited sequence indicate PCR amplicons of TKneo, 5’HA, hPro, hIn2, and hIn6 used in assessing the integrities of hmTert genes in mESC clones. (b) Schematic description of transfection and colony screening strategies. mESCs were co-transfected with the donor BAC and Crispr/Cas9-sgRNA expressing plasmids (pX458-sgRNA1/2) and seeded into 6-cm dishes, followed by puromycin selection. Surviving colonies were individually picked and seeded into 96-well plates, followed by GCV negative selection. The resulting colonies, without cross, were expanded for further characterization by PCR and Southern analyses.