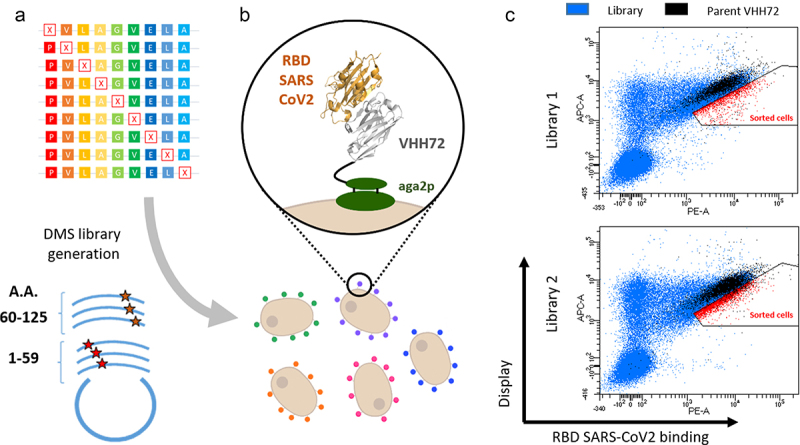
Figure 1.
Deep mutational scanning probing VHH-72 binding to the RBD domain from SARS-CoV2 Spike protein. (a) Two DNA libraries of VHH72 harbouring a single mutation (each corresponding to regions encompassing amino acids 1–59 and 60–125 of the nanobody) were transformed into yeast using gap repair recombination. (b) General principle of functional screening by yeast surface display. Cells are incubated with biotinylated RBD antigen and labelled with secondary reporters before FACS analysis to determine VHH expression and antigen binding. (c) Bivariate flow cytometry analysis of libraries L1 and L2 of yeast cells expressing VHH72 variants on their surface. Cells were double-labelled with biotinylated antigen/Streptavidin–PE (RBD SARS-CoV2 binding) and anti-HA tag antibody coupled to APC (VHH expression). Cells corresponding to clones of the DMS libraries are represented in blue. Libraries were spiked with 10% of clonal cells expressing parental VHH72 along with eGFP protein (represented in black) to discriminate cells with increased antigen binding levels. Selected cells (in red) were sorted and sequenced with illumina deep sequencing.
Alt Text: Deep mutational scanning probing VHH-72 binding to the RBD domain from SARS-CoV2 Spike protein. (a) Scheme illustrating the generation of DMS libraries, their cloning in plasmid and transfection of yeast cells. (b) Scheme representing yeast cells each expressing a mutated VHH72 nanobody. (c) Cytometry dot-plot figures obtained from the analysis of yeast expressing DMS libraries of VHH72.