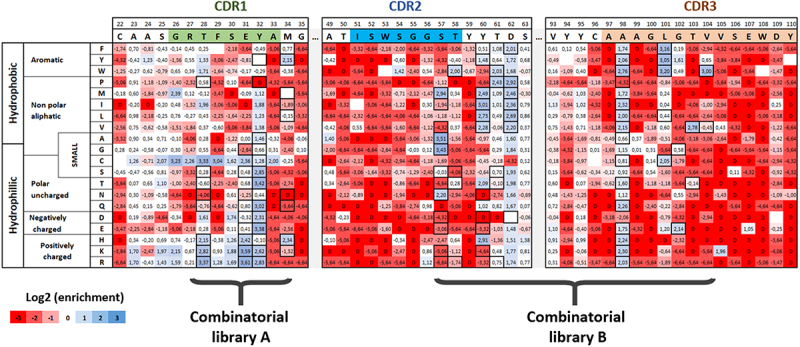
Figure 2.
Paratope mapping of the VHH72 nanobody and design of combinatorial libraries for the selection of clones with improved affinity for SARS-COV-2 RBD antigen. NGS-based heatmap representing enrichment values of each VHH72 single mutant after functional sorting in FACS. Enrichment score is a base 2 log function of enrichment between sorted and unsorted VHH72 yeast populations for a given amino acid substitution. Corresponding table is coloured in blue for enriched mutations and in red for depleted mutations. Black squared substitutions were selected for the design of combinatorial libraries to identify VHH72 variants with improved binding properties. Due to the large generated diversity, two separate libraries were designed (with respective theoretical diversities of 4.14e4 and 2.63e7 clones).
Alt Text: Paratope mapping of the VHH72 nanobody and design of combinatorial libraries for the selection of clones with improved affinity for SARS-COV-2 RBD antigen. Table indicating in for each amino acid position (in column) and each substitution (in rows) the enrichment of the corresponding mutation in the DMS setup. Corresponding table is colored in blue for enriched mutations and in red for depleted mutations. Black squared substitutions were selected for the design of combinatorial libraries to identify VHH72 variants with improved binding properties.