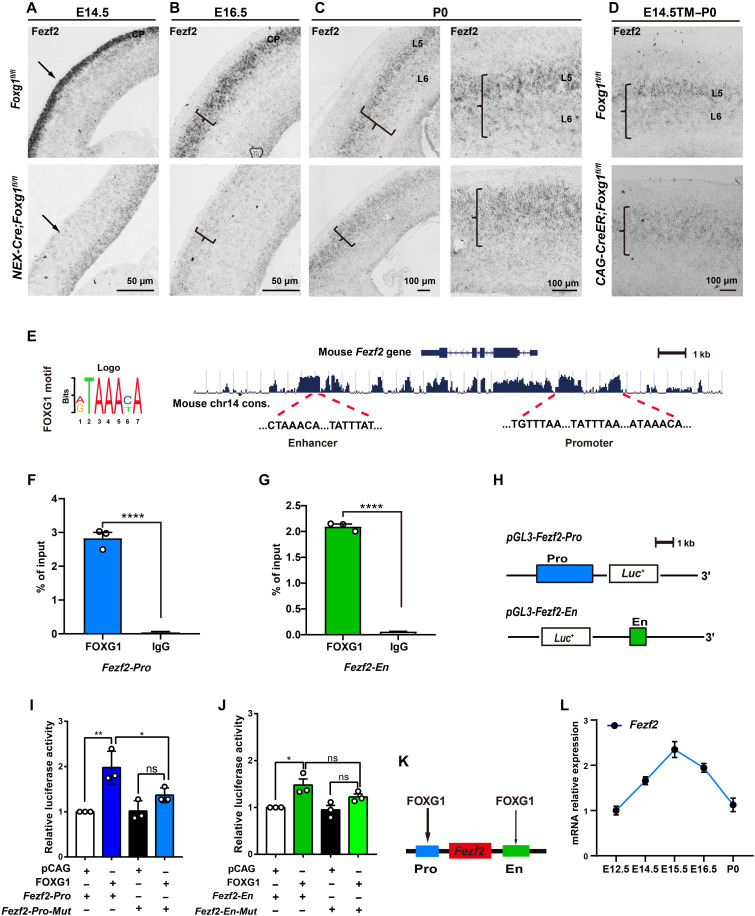
Fig. 4. FOXG1 directly promotes Fezf2 transcription.
(A to D) In situ RNA hybridization showing significantly decreased Fezf2 mRNA level in NEX-Cre;Foxg1 cKO CPs at E14.5 (A) and E16.5 (B). At P0, Fezf2 mRNA expression displayed a SCPNhighCThPNlow pattern in the control but a uniform expression pattern at the NEX-Cre;Foxg1 cKO CPs (C). Similarly, Fezf2 mRNA also displayed a uniform expression pattern in the CAG-CreER;Foxg1 cKO cortex at P0 (D). (E) Motif analyses and prediction of the FOXG1-binding site(s) at the Fezf2 locus. (F and G) ChIP-qPCR of cortex samples at E14.5, showing enriched FOXG1 occupancies at both the promoter and enhancer sites of the Fezf2 locus. (H) Strategy for constructing the vector for the Fezf2 promoter and enhancer for luciferase assays. (I and J) Luciferase assay showing the activation of FOXG1 at the Fezf2 promoter site (I), while showing very weak activation at the Fezf2 enhancer site (J). Deletion of the FOXG1-binding motifs from the Fezf2 promoter reduced the extent of FOXG1-mediated activation of the luciferase reporter. (K) Summary model for FOXG1’s regulation of Fezf2 expression via binding at the Fezf2 promoter and/or enhancer sites. (L) qPCR of cortex samples to monitor Fezf2 mRNA levels over the course of cortical development from E12.5 to P0. Data are presented as means ± SEM; (F and G) unpaired Student’s t test and (I and J) one-way analysis of variance (ANOVA) followed by Tukey-Kramer post hoc test. *P < 0.05, **P < 0.01; ****P < 0.0001.