Fig. 3 |. Bacterial hydroxysteroid dehydrogenases (HSDHs) convert LCA to 3-oxoLCA and isoLCA.
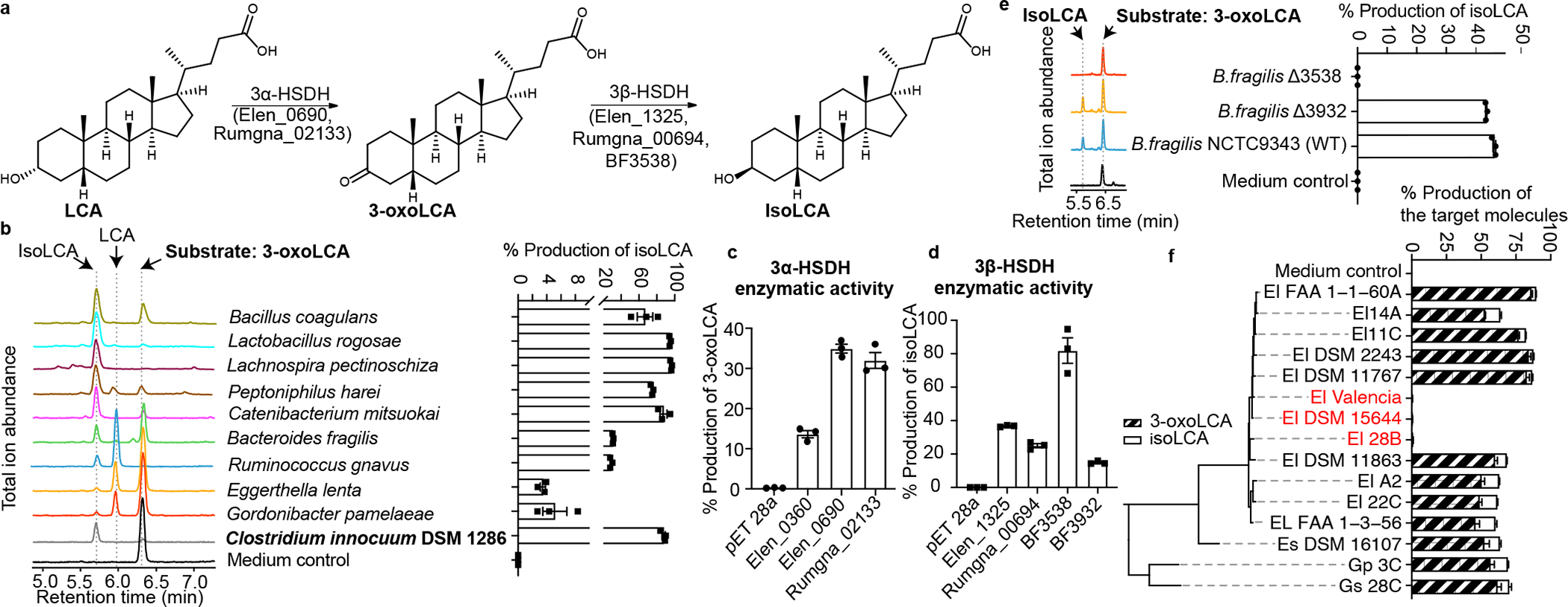
a, Proposed biosynthetic pathway for the conversion of LCA to 3-oxoLCA and isoLCA.
b, Representative UPLC-MS traces (left) and percent production of isoLCA (right) by human bacterial isolates incubated with 3-oxoLCA (100 μM) for 48 hours. TICs are shown. Clostridium innocuum, which encoded 3β-HSDH homologs in HMP2 metagenomes (see Methods; Table S9), was selected for in vitro testing (n = 3 biological replicates per group, data are mean ± SEM, see Table S2).
c, d, Heterologous expression of candidate HSDHs from E. lenta DSM2243, B. fragilis NCTC9343, and R. gnavus ATCC29149 in E. coli. E. coli lysate was incubated with either 100 μM LCA (c) or 100 μM 3-oxoLCA (d) as a substrate. E. coli containing an empty vector was used as a control. Data are reported as percent conversion to product (either 3-oxoLCA or isoLCA) (n = 3 biological replicates per group, data are mean ± SEM, see Extended Data Fig. 4 and Figs. S1 and S10 for protein gels).
e, B. fragilis Δ3538, B. fragilis Δ3932, or the type strain B. fragilis NCTC9343 were incubated with 3-oxoLCA (100 μM) for 48 hours. Representative TIC UPLC-MS traces (left) and percent production of the target molecule isoLCA (right) are shown (n = 3 biological replicates per group, data are mean ± SEM, see Figs. S1 and S11 for DNA gels).
f, Cladogram of E. lenta and related human isolates and their production of 3-oxoLCA and isoLCA (El, E. lenta; Es, Eggerthella sinensis; Gs, Gordonibacter sp., and Gp, Gordonibacter pamelaeae). E. lenta isolates in red (E. lenta 28B, E. lenta DSM15644, E. lenta Valencia) that lack a homolog of Elen_0690 did not synthesize 3-oxoLCA from LCA. All strains were incubated with 100 μM LCA for 48 hours (n = 3 biological replicates per group, data are mean ± SEM).
