Extended Data Fig. 1 |. 3-oxoLCA biosynthetic pathway and microbial diversity from the human screen.
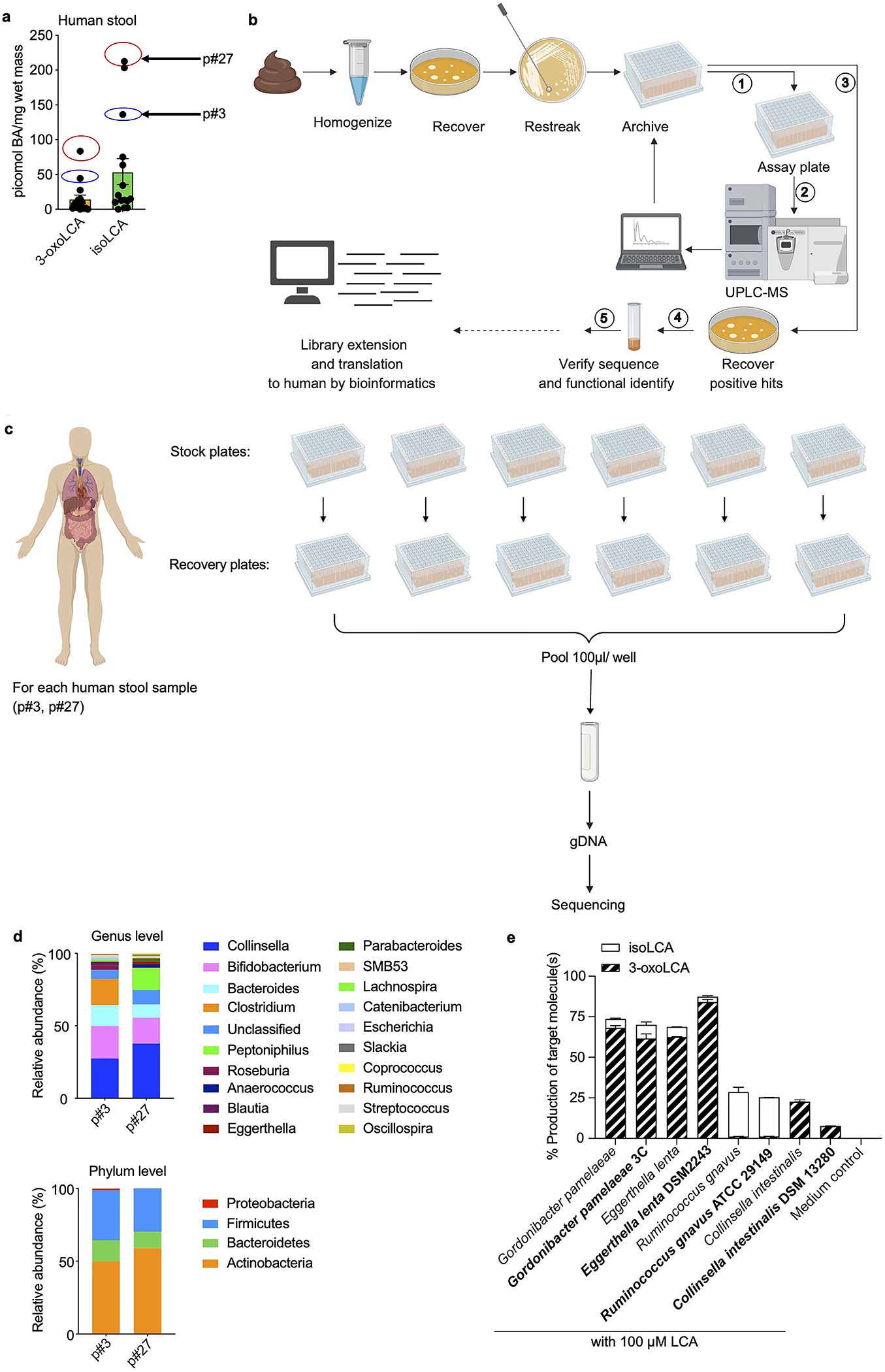
a, Quantification of 3-oxoLCA and isoLCA in stool samples from patients after fecal microbiota transplant (FMT) (n = 15). Stool samples from patient p#3 (3-oxoLCA: 44 picomol/mg, isoLCA: 136 picomol/mg) and patient p#27 (3-oxoLCA: 83 picomol/mg, isoLCA: 213 picomol/mg) were used to screen for 3-oxoLCA producers.
b, Schematic of the screen for bacterial producers of the LCA metabolite 3-oxoLCA from human stool samples. In total, 990 bacterial colonies were isolated, restreaked, and archived from two human stool samples. ① Replicate plates (assay plates) were then used for the screen. ② Individual isolates were incubated anaerobically with LCA (100 μM) (see Fig. 1b) or 3-oxoLCA (100 μM) (see Fig. 2b) for 48 hours. Cultures were harvested, acidified, extracted, and BA metabolites were quantified by UPLC-MS. ③ Positive hits containing 3-oxoLCA were re-selected from the archived stock plates, and recovered on new plates. ④ Activity was verified and each producer species was identified by full-length 16S rRNA sequencing. Finally, bacterial enzymes responsible for the LCA metabolite production were identified (see Fig. 3), and ⑤ corresponding genes were utilized as query sequences in BLASTP searches for novel putative bacterial producers and enzymes.
c, Sample preparation workflow for the determination of cultured bacteria from the human stool sample screen. For each patient, individual isolates were recovered and cultured for 48 hours. These isolates were then pooled together, and genomic DNA was extracted from the pooled pellet. Illumina® MiSeq sequencing on the V3 and V4 hypervariable regions of 16S rRNA was then performed.
d, Genus and phylum-level microbial community composition for each human stool sample.
e, 3-oxoLCA and/or isoLCA production was verified in the type strains of a subset of 3-oxoLCA-producing human isolates (n = 3 biological replicates per group, data are mean ± SEM).
