Abstract
To develop a specific treatment against COVID-19, we investigated silymarin–chitosan nanoparticles (Sil–CNPs) as an antiviral agent against SARS-CoV-2 using in silico and in vitro approaches. Docking of Sil and CNPs was carried out against SARS-CoV-2 spike protein using AutoDock Vina. CNPs and Sil–CNPs were prepared by the ionic gelation method and characterized by TEM, FT-IR, zeta analysis, and the membrane diffusion method to determine the drug release profile. Cytotoxicity was tested on both Vero and Vero E6 cell lines using the MTT assay. Minimum binding energies with spike protein and ACE2 were −6.6, and −8.0 kcal mol−1 for CNPs, and −8.9, and −9.7 kcal mol−1 for Sil, respectively, compared to −6.6 and −8.4 kcal mol−1 respectively for remdesivir (RMV). CNPs and Sil–CNPs were prepared at sizes of 29 nm and 82 nm. The CC50 was 135, 35, and 110 μg mL−1 for CNPs, Sil, and Sil–CNPs, respectively, on Vero E6. The IC50 was determined at concentrations of 0.9, 12 and 0.8 μg mL−1 in virucidal/replication assays for CNPs, Sil, and Sil–CNPs respectively using crystal violet. These results indicate antiviral activity of Sil–CNPs against SARS-CoV-2.
To develop a specific treatment against COVID-19, we investigated silymarin–chitosan nanoparticles (Sil–CNPs) as an antiviral agent against SARS-CoV-2 using in silico and in vitro approaches.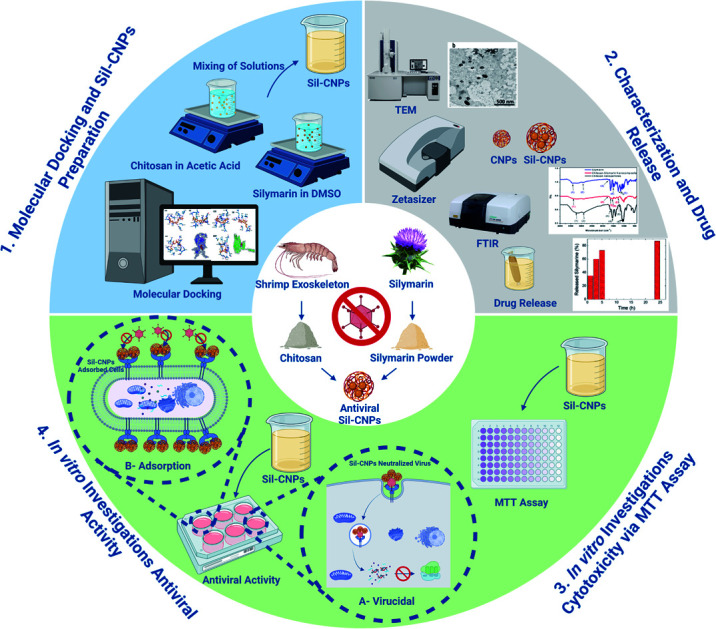
1. Introduction
The current Coronavirus disease-19 (COVID-19) pandemic is exacerbated by the absence of effective therapeutic agents. Patients with acute respiratory syndrome Coronavirus-2 (SARS-CoV-2) infection mostly depend on their own immune defense to control the progress of infection, together with a non-specific treatment protocol to relieve symptoms and improve prognosis. Except for FDA-approved molnupiravir for treatment of non-hospitalized COVID-19 patients,1 no specific antiviral agent is yet available for COVID-19. Therefore, an efficient practical strategy in response to this issue is repurposing drugs with antiviral activity for therapeutic effect.2 In addition, human adenoviruses (HAdV) persist in humans for a long time in latent state and can be reactivated by various factors leading to severe problems specifically in immunosuppressed individuals due to the development of generalized adenovirus infection, also no FDA approved antiviral agents for such viruses.3
Exploring nanotechnology-based approaches for combating COVID-19 might help to overcome the limitations associated with conventional methods of viral disease management.4
Previous studies have reported the successful application of nanostructures in developing virus detection systems and treatment modalities.5,6 Other studies applied nano-based therapy to target Corona viruses, such as SARS-CoV and MERS-CoV.7–10 The first step of the viral infection cycle involves the binding of the virus to the host via cell surface receptors. Blocking the entry of viruses has been found to be a successful anti-viral strategy in many viral infections.11,12
Drug delivery via nanocarriers helps to overcome several challenges associated with the traditional method of antiviral drug administration,13 however, poor bioavailability, susceptibility to in vivo degradation of drug, systemic toxicity, and short half-lifetime in the body are some of the drawbacks associated with this antiviral approach.14
Silymarin (Sil) is a natural flavonolignan substance, known to exhibit anti-oxidative and chemo-protective properties in several viral infections including, dengue, hepatitis B, HIV, influenza, etc.15,16
Recently, they highlighted the rationale for repurposing Sil to treat COVID-19 infections beside its multifaceted beneficial role in a variety of pathophysiological disorders was previously highlighted.17
Moreover, polymeric nanoparticles like chitosan nanoparticles (CNPs) demonstrated interesting properties that allow its usage in antiviral applications, as an efficient and safe drug carrier that helps enhancing the viral therapeutic efficacy and minimizing their adverse effects.17,18 This depends on the biocompatibility, biodegradability, non-immunogenicity, non-toxicity,19 and muco-adhesive properties of chitosan polymer, as it is a natural cationic marine biomaterial.19–21 Moreover, it is derived from the exoskeleton of crustaceans, insects and fungal cell walls in the form of chitin and then treated by alkaline deacetylation. It is considered as an ideal biopolymer20 and is approved by FDA for using in biomedical applications.19
Our previous work that was based on encapsulating curcumin into CNPs22 has demonstrated an increase in antiviral activity against hepatitis C genotype 4 (HCV-4a) proved on the molecular and protein levels.
In the current study, we proposed a similar approach to develop an antiviral agent against SARS-CoV-2 and adenovirus type 5 (ADV-5), based on encapsulating Sil into CNPs as an efficient and highly specific drug delivery system. This can increase antiviral activity against both viruses, thus controlling their transmission. To achieve such approach, in silico study was performed for Sil and CNPs against spike protein of SARS-CoV-2 and against capsid protein (Hexon) of ADV-5, then a panel of chemical and biological investigations was performed to evaluate antiviral activity of nanoparticles and its composite before proceeding to in vivo trial.
2. Materials and methods
2.1. Materials
Silymarin (Sil) was purchased from Gilvar Inc., Markham, ON, Canada. Phosphate-buffered saline (PBS) tablets were purchased from Fisher Scientific, Loughborough, UK. Chitosan (low molecular weight), tripolyphosphate (TPP), ethanol, methanol, ethyl acetate, glutaraldehyde, and isopropanol were purchased from Sigma Aldrich, St Louis, MO, USA. MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] reagent was purchased from Serva, Heidelberg, Germany. Dulbecco's modified Eagle's medium (DMEM), penicillin/streptomycin, and fetal bovine serum (FBS) were purchased from Gibco, Merelbeke, Belgium. Vero cell line was obtained from Vaccination and Sera Collection Organization (VACSERA), Agouza, Giza, Egypt. The Vero E6 cells were kindly provided from the cell culture collection of Nawah Scientific, Al-Asmarat, Almokattam, Cairo, Egypt. Human adeno virus type-5 (ADV-5, VR-5, ATCC).
2.2. Methods
2.2.1. In silico docking study
In the current study, structure of Hexon proteins of the HAdV were retrieved from the protein data bank PDB ID 6CGV and protein structures for ACE2 and SARS-CoV-2 spike proteins were retrieved from PDB ID 6m17, while the structures of chitosan polymer, Sil and remdesivir (RMV) molecules were built using the free software Avogadro. All pdbqt files were prepared using Autodock tools and docking was performed by AutoDock Vina. The Protein–Ligand Interaction Profiler (PLIP) analysis was used to investigate different interactions inside the predicted binding pockets between ligand and protein.
2.2.2. Preparation of CNPs and Sil–CNPs
CNPs were prepared by dissolving (1 g) chitosan bulk in 100 mL (1% acetic acid solution in DI H2O) until the solution was transparent. The chitosan solution was then neutralized by the addition of the negatively charged tripolyphosphate (TPP) solution (0.2 g of TPP dissolved in 100 mL DI H2O) under magnetic stirring (700 rpm) for 2 hour (h) at room temperature. The obtained nanoparticles were then washed several times with distilled water followed by centrifugation at 15 000 rpm to remove the residual acetic acid and other impurities.
Sil–CNPs was prepared by dissolving Sil powder (0.581 g) in 5 mL of DMSO to prepare a 0.241 M solution. The Sil solution was then added to the chitosan solution and mixed with continuous stirring for 1 h. The preparation steps were completed as in preparation of CNPs.
2.2.3. Characterization of CNPs and Sil–CNPs
The morphologies of CNPs and Sil–CNPs were investigated with a Transmission Electron Microscope (Joel JEM-2100) operated at 200 kV. The samples were dispersed in ethanol, then a copper carbon coated grid was immersed in the dispersed sample solution and left to dry before imaging.
The chemical structures of nanomaterials were analyzed with FT-IR (Bruker Vertex 70, Germany) with the IR fingerprints recorded between 4000-400 cm−1 using transmittance modes.23,24
2.2.4. Encapsulation efficiency and loading capacity
The encapsulation efficiency of Sil in CNPs was determined by placing 25 mg of Sil–CNPs in 60 mL of distilled water. The resulting solution was stirred and centrifuged at 37 564 rcf for 15 minutes (min), then the supernatant was collected. The amount of drug within the supernatant was estimated using a spectrophotometer (Agilent Technologies, Cary series UV-Vis-NIR, Korea) at an absorption maximum of 260 nm, which corresponds to the peak absorption of Sil. The entrapment efficiency was calculated using the following equation:25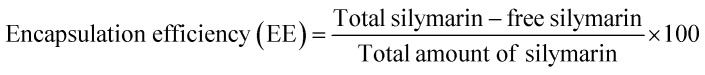
The loading capacity is the ratio of weight of encapsulated material to the weight of nanoparticles, and it was calculated according to the following equation: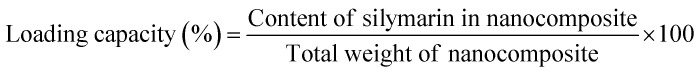
2.2.5. In vitro drug release
The Sil release profile from CNPs was determined using a membrane diffusion method. Briefly, 25 mg of Sil–CNPs were placed in a dialysis bag containing 10 mL distilled water (12–14 kDa MWCO, Visking dialysis tubing, SERVA, Germany). Then, the dialysis bag was inserted and fully immersed in 10 mL distilled water as a release medium at 37 °C and pH 7 with gentle stirring. At various time intervals, the concentration of Sil in samples was measured using a spectrophotometer at 280 nm.
2.2.6. Cell culture
Vero and Vero E6 cell lines were sub-cultured and maintained in DMEM containing FBS (10%), and antibiotics (100 IU mL−1 penicillin/streptomycin, Lonza, Belgium). Cells were propagated as a monolayer culture at 37 °C under a humidified atmosphere of 5% CO2. Finally, cells were routinely sub-cultured by trypsinization (trypsin/versine, 0.05%, Lonza).
2.2.6.1. Cytotoxicity evaluation using MTT colorimetric assay on Vero and Vero E6 cell lines
We used a modified method utilizing MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) dye that is based on the reduction of the dye by mitochondrial dehydrogenases of metabolically active cells into insoluble formazan crystals. Briefly, cell monolayers were cultured for 48 h before exposure to MTT solution in PBS (5 mg mL−1) and incubated for 90 min. The formation of formazan crystals was visually confirmed using phase contrast microscopy. DMSO (100 μL per well) was added to dissolve the formazan crystals with shaking for 10 min. The absorbance was measured at 540 nm against blanks (media only) on a microplate reader (CLARIOstar plus, BMG LabTech, Germany). Cell survival index was calculated by comparing the optical density (OD) values of the DMSO control wells with those of the samples and expressed as % viability to the control. The dose–response experiment was performed on samples producing ≥50% loss of cell viability using five serial two-fold dilutions (200, 100, 50, 25, 12.5, and 6.25 μg mL−1) of the sample. Where applicable, CC50 values (concentration of sample causing 50% loss of cell viability of the vehicle control) were calculated using the dose response curve fit to non-linear regression correlation using GraphPad Prism® V6.0 software.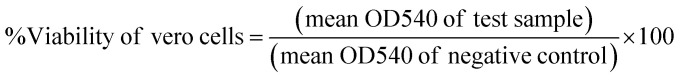
2.2.6.2. Morphological examination
Morphological changes were tested after treatment of Vero and Vero E6 with all tested material after 24 h under an inverted microscope to confirm MTT results.
2.2.7. Cellular uptake using STEM technique
A previously cultured Vero cells (3 × 106) in 75 cm2 flask (Corning, USA) were treated with 100 μg mL−1 of Sil–CNPs for 24 h. Cells were then washed with PBS buffer, fixed with 2% glutaraldehyde for 2 h, washed twice with PBS and finally fixed in 1% OsO4 for 1 h. Following agarose (1.5%) enrobing, Spurr's resin embedding, and ultrathin (50 nm) sectioning, the samples were stained with 2% aqueous uranyl acetate and 25 mg mL−1 lead citrate and imaged with a STEM detector attached to FESEM (Quattro S, Thermo Scientific).26
2.2.8. Antiviral assay against human adenovirus-5 (ADV-5) using real-time PCR assay
This step included 3 stages:
2.2.8.1. Infectivity assay
Adenovirus type 5 ADV-5 (VR-5) was propagated in Vero cells and investigated daily under an inverted microscope until 80–90% of CPE and cell lysis was observed (this was obtained after 48 h of infection). The virus culture was subjected to three times of freezing and thawing for determination of viral load using quantitative real-time PCR assay.27 Standard curve was constructed using ten-fold serial dilutions (10 to 106 copies per mL) of standard adenovirus type-5 (VR-5) control.
2.2.8.2. Viral titration
Before evaluating the tested materials for its antiviral activity, we titrated the virus using MTT assay and the IC50 (dilution of the virus that can kill 50% of the cells) was used in the antiviral assay. This method was performed according to previously published protocol.28
2.2.8.3. Antiviral mechanism
The non-toxic doses of the tested material were used at concentration of 100 μg for both CNPs and its composite and 20 μg mL−1 for Sil. Three antiviral mechanisms were performed including:
2.2.8.3.1. Adsorption mechanism
A 6-well plate was seeded with 5 × 105 cells per mL and incubated at standard condition, 37 °C and 5% CO2. After 24 h, the cells were treated with non-toxic concentration of our materials (as confirmed by cytotoxicity assays and morphological examination). After 24 h, the cells were infected with 100 000 copies per mL of adenovirus stock and incubated for 24 h at standard condition. Controls were used in each assay: untreated well for cell control; untreated infected well for viral control. The plate was then subjected for quantitative real-time PCR assay to measure viral load.
2.2.8.3.2. Viral replication mechanism
A 6-well plate was seeded with 5 × 105 cells per mL and incubated at standard condition. After 24 h, the cells were infected with 1000 copies per mL of adenovirus stock and incubated for 24 h at standard condition, then the cells were washed and treated with non-toxic concentration of our materials and incubated for another 24 h at standard condition. Controls were run in each assay: untreated well for cell control; infected well untreated for viral control. The plate was then subjected for quantitative real-time PCR assay to measure viral load.
2.2.8.3.3. Virucidal mechanism
This method was performed to investigate ability of the tested materials to neutralize and inactivate viral particles and then block its ability to enter the cells. A 6-well plate was seeded with 5 × 105 cells per mL and incubated at standard condition. After 24 h, the cells were treated with non-toxic concentration of our materials after its incubation with virus (1 : 100 000 copies per mL) at 4 °C for 1 h, then continue incubation for another 24 h at standard condition. The plate was then subjected for quantitative real-time PCR assay to measure viral load. Based on the previous effective antiviral mechanisms against ADV-5, our materials were subjected to the most effective antiviral activity mechanism.
2.2.9. Antiviral activity against SARS-CoV-2 on Vero E6
2.2.9.1. Antiviral activity and viral titration using crystal violet assay
In 96-well tissue culture plates, 2.4 × 104 Vero-E6 cells were distributed in each well and incubated overnight at a humidified 37 °C incubator under 5% CO2 condition. The cell monolayers were then washed once with 1× PBS. In cell adsorption assay, the Vero-E6 cells were treated with serial dilution of compounds for 24 h before infection. 24 h later, compounds were removed, and cells were subjected to infection with 100 TCID50 of SARS-CoV-2 virus for 72 h in a total volume of 200 μL per well. Untreated cells infected with virus represent virus control, however cells that are not treated and not infected are cell control. 72 h post-infection, plates were treated by 10% formaldehyde and incubated for 2–4 h. Cell monolayers are then washed and stained with 0.5% crystal violet and incubated for 10–30 min. Plates were further washed, dried and the crystal violet stain was dissolved with 200 μL methanol. The intensity optical density of the dissolved crystal violet is measured at 570 nm.
In virus replication assay, cells were not treated with compounds, but instead, 100 TCID50 of SARS-CoV-2 “NRC-03-nhCoV virus”,29,30 was incubated with serial diluted compounds and kept at 37 °C for 1 h. Then, cell monolayers were treated with 100 μL of virus/compound mix and co-incubated at 37 °C. After 1 h, another 100 μL of infection medium were added and the plate was incubated for 72 h. Following incubation at 37 °C in 5% CO2 incubator for 72 h, the cells were fixed with 100 μL of 10% paraformaldehyde for 20 min and stained with 0.5% crystal violet in distilled water for 15 min at room temperature. The crystal violet dye was then dissolved using 100 μL absolute methanol per well and the optical density of the color is measured at 570 nm using Anthos Zenyth 200 rt plate reader (Anthos Labtec Instruments, Heerhugowaard, Netherlands). The IC50 of the compound is that concentration required to reduce the virus-induced CPE by 50%, relative to the virus control.
2.2.9.2. Standard curve for quantitative measurement of SARS-CoV-2 using real-time RT-PCR assay
RNA extraction was performed using QIAamp viral RNA extraction kit (Qiagen, Valencia, USA), according to the manufacturer's instructions. The RNA extract was used in real-time PCR assay using Vitro S.A One-Step RT Kit (Sevilla, Spain) with a total reaction volume of 20 μL. The reaction consisted of four stages: stage 1 at 25 °C for 5 min, stage 2 at 50 °C for 20 min, stage 3 at 95 °C for 5 min followed by stage 4 of repeated cycles (45) that consisted of 95 °C for 30 s then 60 °C for 1 min. Standard curves were constructed using Serial dilutions from 106 copies per reaction to 10 copies per reaction of synthetic fragments of the N gene in the FAM channel and E gene in the ROX channel and the both genes were detected at 10 copies per reaction.
2.2.9.3. Antiviral assay of SARS-CoV-2 using real-time PCR assay
The antiviral activity was performed in a six-well plate in the same procedure performed for adenovirus but instead hCoV-19/Egypt/NRC-3/2020 isolate was used at 0.01 multiplicity of infection (MOI) via two mechanisms adsorption and replication. Then cells were subjected to three times freezing and thawing, viral extraction and real-time RT PCR assay for quantification of viral load per mL.
3. Results and discussion
3.1. Molecular docking calculation of CNPs, Sil and remdesivir (RMV) against capsid protein Hexon, SARS-CoV-2 Spike protein and human ACE2 protein
Sil showed the highest binding affinity (∼−9.7 kcal mol−1) against ACE2 protein in the predicted binding pocket, while CNPs and RMV molecules bounded with comparable energies of −8.9 and −8.4 kcal mol−1, respectively. For SARS-CoV-2 spike protein, the binding energies for Sil was −8.0 kcal mol−1 in comparison to that of CNPs and RMV were equal (−6.6 kcal mol−1 for both). Molecular docking calculations showed a favorable interaction of ligands against Hexon protein (PDB ID 6CGV). RMV binding to Hexon protein was −8.2 kcal mol−1 and was thus the highest binding energy compared to other ligands with the same protein (Table 1).
Binding energies of silymarin, chitosan nanoparticles, and remdesivir against 1st binding pose predicted by Autodock Vina in kcal mol−1 against SARS-CoV-2 Spike protein, human ACE2, and Hexon of Adenovirus type-5.
| Sil | CNPs | RMV | |
|---|---|---|---|
| SARS-CoV-2 spike protein | −8.0 | −6.6 | −6.6 |
| ACE2 | −9.7 | −8.9 | −8.4 |
| Hexon | −7.8 | −6.8 | −8.2 |
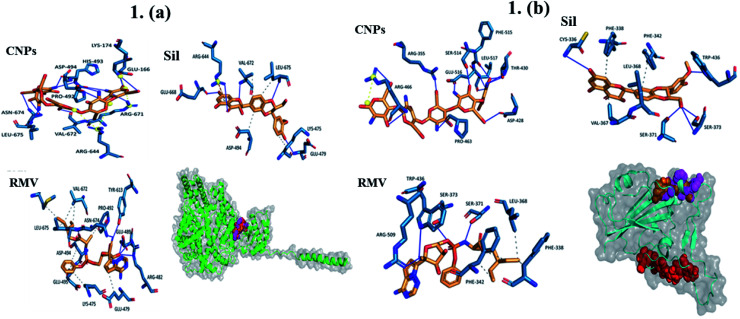
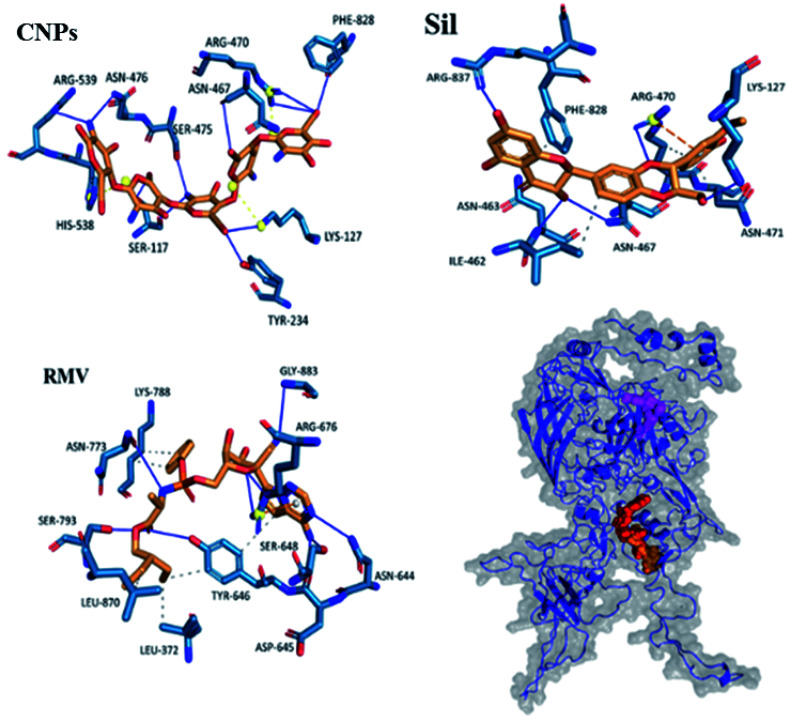
Interactions of the three ligands in the predicted binding pockets were analyzed by the Protein–Ligand Interaction Profiler (PLIP) analysis online tool (Fig. 1 and 2). Interactions of CNPs with Hexon, ACE2 and SARS-CoV-2 spike proteins was dominated by hydrogen bond interactions. Moreover, there were few salt bridges formed between charged amino acids and CNPs polymer. While for both of Sil and RMV, binding was mainly, due to both of hydrogen bond and hydrophobic interactions. Moreover, phenol groups in both of Sil and RMV induced π–cation interactions with positive charge center of the guanidinium groups in ARG residues in both of predicted binding pockets of the Hexon and ACE2 proteins. Residues interacting with ligands (CNPs, Sil, and RMV) in the predicted Hexon, ACE2, and Spike protein binding pockets are presented in (Table S1†).
Fig. 1. (a) Interaction of CNPs, Sil, and RMV with ACE2 (orange, red, and magenta, respectively.) ____ Hydrogen interactions, - - - - hydrophobic interactions, yellow spheres are charge centers, - - - - salt bridges, and - - - - is π–cation Interactions. (b) Interaction of CNPs, Sil, and RMV with SARS-CoV-2 (orange, red, and magenta, respectively). ____ Hydrogen interactions, - - - - hydrophobic interactions, yellow spheres are charge centers, - - - - salt bridges, and - - - - is π–cation interactions.
Fig. 2. Interaction of CNPs, Sil, and RMV with Hexon protein (orange, red, and magenta, respectively). ____ hydrogen interactions, - - - - hydrophobic interactions, yellow spheres are charge centers, - - - - salt bridges, and - - - - is π–cation interactions.
Results showed that interactions of CNPs with adenovirus Hexon, ACE2 and SARS-CoV-2 spike proteins are mainly hydrogen bond interactions. Moreover, there is a presence of few salt bridges formed between charged amino acids and CNPs. While for both of Sil and RMV, binding was shown to be mainly due to both hydrogen bond and hydrophobic interactions. Moreover, phenol groups in both of Sil and RMV induced π–cation interactions with positive charge center of the guanidinium groups in ARG residues in both predicted binding pockets of the Hexon and ACE2 proteins. Such promising results encouraged designing and preparation of Sil–CNPs to increase antiviral activity and continuing in vitro study in susceptible cell culture system.31
Sil extract has some limiting factors to be used as drug such as its lower bioavailability, lower solubility in water, and poor absorption in the gastrointestinal tract. Therefore, several attempts have been made to improve its solubility, and bioavailability.32
In continuation of our previous report, by studying CNPs was used as drug carrier for curcumin and proved its multi target antiviral agent against HCV-4a entry and replication.17 Hence, in this study we explored the antiviral activity of CNPs, Sil–CNPs, and Sil against ADV-5 and SARS-CoV-2 in cell culture system.
3.2. Preparation and characterization of chitosan nanoparticles (CNPs) and silymarin–chitosan nanocomposite (Sil–CNPs)
CNPs and Sil–CNPs were synthesized using ionic gelation method.33 CNPs and Sil–CNPs were synthesized using the ionic gelation method and were evaluated for their efficiency against SARS-CoV-2.
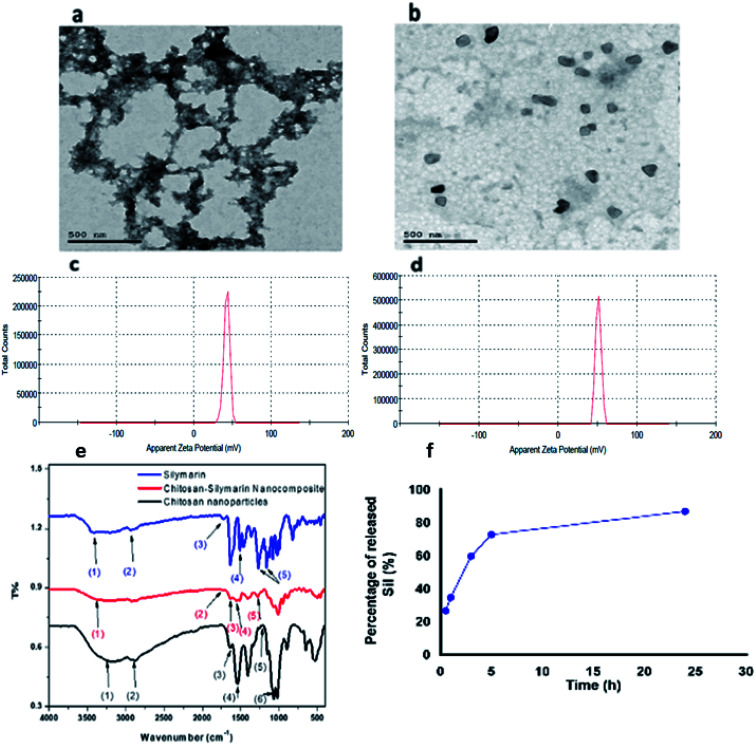
The prepared materials were subjected to several characterization procedures to confirm the binding and study their binding properties (Fig. 3a). shows Transmission electron microscope (TEM) images, revealing CNPs that have an average size of about 29 nm, and that some agglomerated particles are observed with no clearly defined shape (irregular), while Sil–CNPs nanocomposite showed nearly spherical shape with an average particle size of 82 nm, as shown in (Fig. 3b), which was smaller than that reported by Aboshanab et al.34 which was 100 nm, also the particle shape reported in their study was agglomerated particles with rounded shape which is different from that reported in our current study.
Fig. 3. Characterization of CNPs and Sil–CNPs composite: (a and b) TEM images, (c and d) zeta potential, (e) FT-IR, and (f) drug release.
Fig. 3c shows the Fourier-transform infrared (FT-IR) spectra of Sil, CNPs, and the composite. The wavenumbers of the corresponding vibrational modes of Sil, CNPs, and Sil–CNPs are shown in (Table 2).
Wavenumbers corresponding to the vibrational modes of Sil, CNPs, and Sil–CNPs.
| No. | Wavenumber (cm−1) | Vibrational modes |
|---|---|---|
| Chitosan/TPP | ||
| 1 | 3190 | O–H & N–H stretch |
| 2 | 2870 | C–H stretch |
| 3 | 1630 | C O stretch acetyl GP |
| 4 | 1540 | N–H bending |
| 5 | 1212 | P O stretching |
| 6 | 1066 | C–O asymmetric stretch |
| Sil | ||
| 1 | 3425 | Phenolic (OH) stretching |
| 2 | 2870 | C–H stretching |
| 3 | 1732 | Ketonic (C O) |
| 4 | 1510 | Ring stretching C C |
| 5 | 1162, 1268 | C–O–C stretching |
| Sil–CNPs | ||
| 1 | 3289 | OH stretching |
| 2 | 1732 | Ketonic (C O) |
| 3 | 1540 | N–H bending |
| 4 | 1510 | Ring stretching C C |
| 5 | 1268 | C–O–C stretching |
It was found that the Sil conjugated to CNPs via hydrogen bond and electrostatic attraction, as revealed by the FT-IR. The spectra of Sil showed peaks at 3425 cm−1 corresponding to phenolic (OH) vibrations, 1732 cm−1 corresponding to (C O) ketonic, 1510 cm−1 attributed to the symmetric aromatic ring stretching vibration (C C ring), 1162 cm−1 and 1268 cm−1 corresponding to C–O–C stretching vibration, and 2870 cm−1 attributed to C–H stretching vibration.35
The FT-IR analysis of the CNPs showed peak at 1212 cm−1 which is assigned to the stretching vibration of P O that is introduced during cross linkage reaction between chitosan and TPP. Peaks at 3190 cm−1, 1630 cm−1 and 1540 cm−1 were assigned to stretching vibrations of OH, C O, N–H bending of amide II, respectively. Moreover, the appearance of characteristic peaks of CNPs at 2870 cm−1 and 1066 cm−1 were attributed to the C–H and C–O groups' stretching vibrations.36 After loading Sil into CNPs, a broadening in OH group was observed, as well as a shift from 3425 cm−1 into 3289 cm−1, indicating the formation of hydrogen bond between Sil and CNPs. The peak at 1540 cm−1 that attributed to N–H bending of amide II in CNPs appeared, but with lower intensity, indicating electrostatic attraction between Sil and CNPs. These indicate that Sil was conjugated to CNPs via hydrogen bond and electrostatic attraction. In addition to the appearance of the Sil characteristic peaks at 1732 cm−1, 1510 cm−1, 1268 cm−1 that attributed to ketonic (C O), ring stretching C C, and C–O–C stretching respectively at the FT-IR spectra of the composite, which confirms loading of Sil into CNPs. The schematic diagram of the chemical structure Sil–CNPs is presented in Fig. S1.†
3.3. Entrapment efficiency and loading capacity
The entrapment efficiency and loading capacity of Sil–CNPs are estimated to be ≈0.263% and 0.1%, respectively. It is recommended to increase chitosan : TPP ratio to subsequently increase the entrapment efficiency of the drug into the nanocomposites.
3.4. Drug release
Interestingly, the release of Sil initially occurred rapidly, about 35% of Sil was released within the first hour of release process as a “burst release effect”, which is the result of fast release of the loaded Sil to the surface or near to the surface of CNPs. This was then followed by a slow Sil release rate, until reaching 87% as an accumulated released Sil after 24 h. Thereafter, the release of Sil from the nanocomposite proceeded in a biphasic release profile (Fig. 3d). Such biphasic rate explained safety of nanocomposite compared to Sil alone where CC50 of nanocomposite reached to >100 μg mL−1 compared to 60 μg mL−1 for Sil alone. In contrast, Gupta and coworkers37 observed that encapsulating of Sil into CNPs provided a sustained release behavior of Sil from the prepared Sil–CNPs. Moreover, they reported that only 52.6% of the total loaded Sil was released from CNPs after 24 h, while our results demonstrated a somewhat higher release of Sil reaching 87% after 24 h.37
3.5. Interaction of CNPs and Sil–CNPs with Vero cells
3.5.1. Cell culture and MTT colorimetric assay
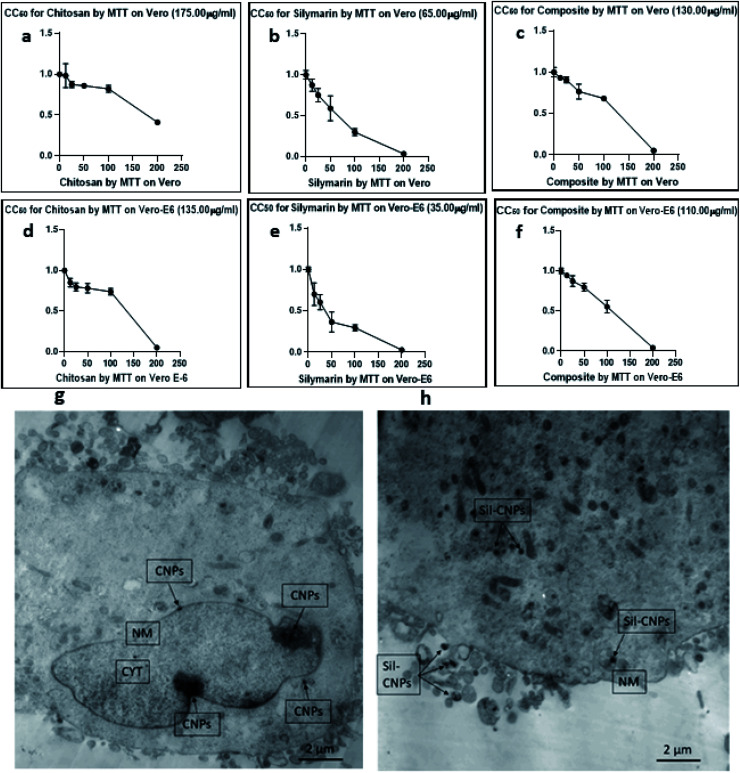
The cytotoxic effect of various concentrations (6.25, 12.5, 25, 50, 100, and 200 μg mL−1) of Sil, CNPs and Sil–CNPs on the viability of Vero and Vero E6 cells, was determined using MTT colorimetric assay after 48 h of exposure. Both CNPs and Sil–CNPs did not reveal cytotoxic effect on the cells at concentrations up to 100 μg mL−1 during the 48 h period making them good candidates for investigating their antiviral activity.
CC50s of CNPs, Sil, and Sil–CNPs were determined at 175, 65, and 130 μg mL−1 on Vero cells and at 135, 35, and 110 μg mL−1 on Vero E6 cells. These results are demonstrated in (Fig. 4e and f).
Fig. 4. (a–f) Cytotoxicity tests of (a) CNPs, (b) Sil, (c) Sil–CNPs composite on Vero cells, (d–f) cytotoxicity of (d) CNPs, (e) Sil, and (f) Sil–CNPs composite on Vero E6 cells. Cellular uptake of (g) CNPs, and (h) Sil–CNPs using TEM after 24 h cell exposure. *CNPs: chitosan nanoparticles, Sil–CNPs: silymarin–chitosan nanoparticles, NM: nuclear membrane, and CYT: cytoplasm. These figures (a–f) were replaced by CC50 instead of IC50.
Our results agrees with that of our previous report which demonstrated that the cytotoxic effect of our prepared chitosan–curcumin nanocomposite was lower than that of free curcumin.22
3.5.2. Cellular uptake
Cellular uptake for CNPs and its nanocomposite (Sil–CNPs) was performed on Vero cells at concentration of 100 μg mL−1. This concentration was used to evaluate localization of nanocomposite inside cellular compartments using TEM which demonstrated its internalization into cellular organelles like cytoplasm and nucleus (Fig. 4g and h).
3.6. Antiviral assay against ADV-5 and SARS-CoV-2
3.6.1. Antiviral assay against ADV-5 using real-time polymerase chain reaction (PCR) assay
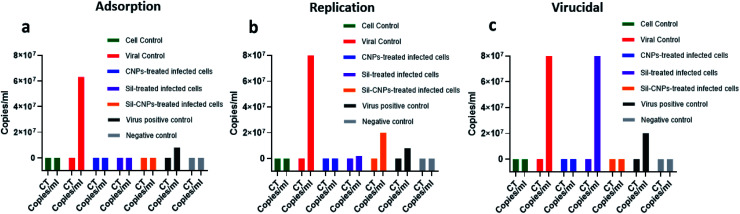
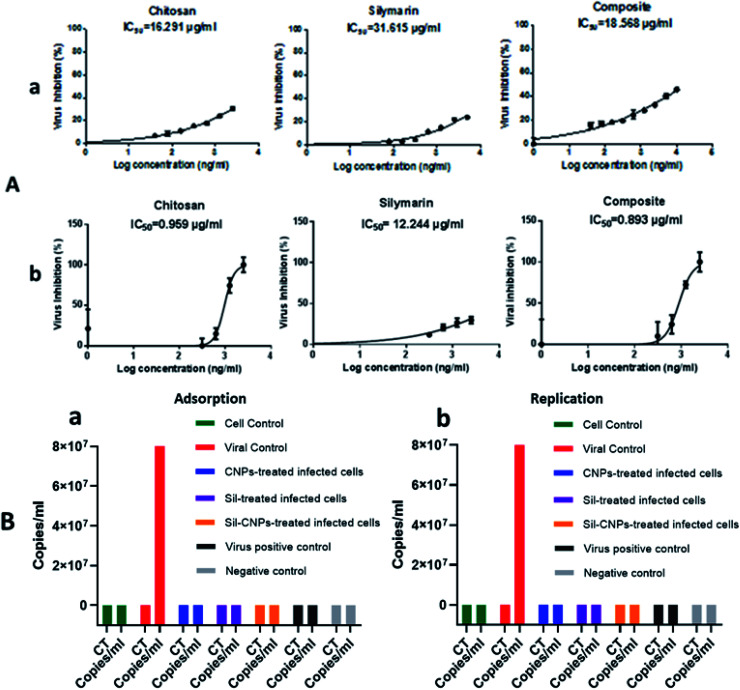
Quantitative measurement of antiviral activity against ADV-5 was determined for our tested materials using real-time assay in the three antiviral mechanisms included adsorption, replication and virucidal. Results showed that all our compounds showed antiviral activity via adsorption and virucidal mechanisms for Sil, CNPs, and Sil–CNPs at concentrations of (20, 100, and 100 μg mL−1, respectively) as evidenced by undetected level of viral copies/mL (Fig. 5), except Sil alone didn't show virucidal antiviral activity as shown in (Fig. 5c). This indicates direct effect of our nanoparticles and its nanocomposite in altering structure of adenovirus, then reducing its infectivity and hence couldn't be detected by quantitative PCR assay. It has been reported that amino groups (NH2) and hydroxyl groups (OH) are functional groups that mediate the antiviral activity of CNPs.38 The molecular weight and degree of de-acetylation of CNPs also play an important role in defining the biological activity.
Fig. 5. Antiviral activity against ADV-5 using real-time PCR assay using (a) adsorption, (b) replication, and (c) virucidal mechanisms.
In addition, positive charges of nanoparticles increase its binding affinity to negative charges on cell membrane. One study suggested that aminoethyl-modified chitosan exerted its antiviral activity by stimulation of the immune response and demonstrated its application for vaccination against several viral infections.18
Regarding antiviral replication mechanism, CNPs decreased viral titer by only 10 folds, but Sil decreased it by 100 folds compared to viral control (Fig. 5). Therefore, it is quite relevant to investigate all possible antiviral mechanisms for any query tested antiviral material.
It has been reported that possible mechanisms of antiviral activity of nanoparticles involved (1) in activation of the virus directly or (2) indirectly by preventing viral attachment to host cells then blocking viral entry and hence its replication, this depends on form and type of applied nanoparticles.39
Moreover, we reached IC50 of composite after using 0.2% of Sil which means we enhanced antiviral activity of Sil after its conjugation to CNPs.
This was also previously supported by the work of Lysenko et al., who proposed that one of the main and direct mechanisms of nanoparticle-mediated antiviral activity is linked to local-field action against the receptors at the virus surface changing the membrane potential.39,40
These results are also supported by in silico study showing high binding affinity of both CNPs and Sil against Hexon.
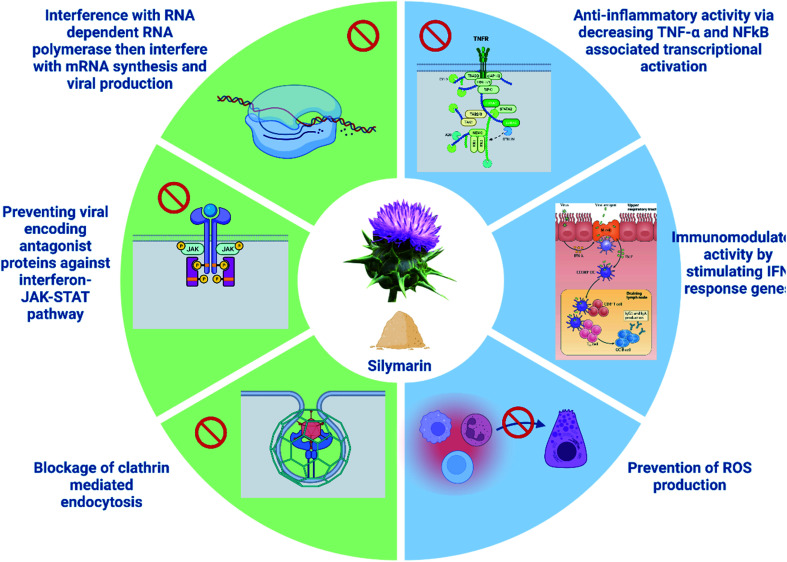
Moreover, previously, it has been reported that Sil prevents replication of influenza A virus, via several mechanisms. It inhibits late mRNA synthesis as well as inducing JAK-STAT antiviral signaling pathway, which leads to production of gamma interferon that exerts its immunomodulatory function and activates cellular processes (such as development of WBCs, gamma interferon production, etc.).16,41 The antiviral mechanisms are demonstrated in (Fig. 6).
Fig. 6. Antiviral mechanisms/pathways exhibited by silymarin (created by Biorender).
Therefore, in the current study we assessed the antiviral activity of CNPs alone and after conjugation to Sil which is one of the promising antiviral candidate.16
3.6.2. Antiviral activity against SARS-CoV-2 on Vero E6
3.6.2.1. Antiviral activity and viral titration using crystal violet assay
Vero-E6 cells were pretreated with nontoxic concentrations of each compound (100, 20, and 100 μg mL−1 for CNPs, Sil, and nanocomposite, respectively), where angiotensin-converting enzyme 2 (ACE2) was previously identified in Vero E6 cells as a receptor for SARS-CoV42 followed by viral infection at 0.1 multiplicity of infection (MOI) of NRC-03-nhCoV resulted in IC50 at concentrations of 16, 31 and 18 μg mL−1 for CNPs, Sil, and Sil–CNPs, respectively (Fig. 7A). Whereas, in virucidal/replication antiviral assay, IC50 was determined at 0.9, 12, and 0.8 μg mL−1 respectively for the same material (Fig. 7B). This indicates their ability to inhibit or compete with viral binding to the host cell-surface receptor, i.e. ACE2 receptor, but results in virucidal/replication mechanism were reached at lower concentrations than that in the adsorption mechanism.
Fig. 7. Antiviral activity against SARS-CoV-2 on Vero E6 cell line using (A) crystal violet assay (a) adsorption mechanism & (b) replication/virucidal mechanism, and (B) real-time PCR assay (a) adsorption mechanism & (b) replication/virucidal mechanism.
3.6.2.2. Antiviral activity using real time RT-PCR assay
Regarding, results of real-time RT PCR assay showed that antiviral activity was achieved only in the adsorption mechanism. This indicates that the main target of antiviral activity of our prepared nanoparticles and its nanocomposite (Sil–CNPs) against SARS-CoV-2 was through interference with viral attachment by blocking ACE2 receptors, thus preventing viral entry which is expected based on previously published report.41
But what was of interest here is the inability of Sil alone to prevent viral infectivity, as we still could detect viral copies/mL using real-time RT PCR assay. However, RT PCR assay is based on amplification of any segment of the target, therefore, it cannot rule out antiviral activity of the tested material on other viral proteins. In addition, our results were supported by in silico analysis which showed high binding affinity of CNPs against ACE2 receptor which makes it a quiet promising candidate for preventing viral entry, especially its mucoadhesive property healing in its application as intranasal vaccine targeting mucosal pathway inducing local humoral and cellular immune responses.43,44
4. Conclusion
In silico study showed high binding energies of our tested agents, especially SARS-CoV-2. CNPs showed a promising antiviral agent against ADV-5 and SARS-CoV-2 in vitro and its conjugation to Sil improved its bioavailability and physicochemical properties. The increased antiviral activity might be via blocking viral host receptor ACE2, thus preventing viral attachment and its entry to the cells. These results were verified using real-time PCR assay. These results may need further investigations using TEM to confirm blockage of receptor during viral infection and some metabolic profile, before proceeding to in vivo study probably in the form of intranasal drug delivery to inhibit viral entry. The future should focus on development of antiviral nano-based therapy as an effective way to control viral causing diseases.
Author contributions
S. A. L.: PI of ASRT 6901, funding acquisition, project administration, conceptualization, editing, supervision. A. A. S.: preparation and characterization of chitosan, silymarin, and composite nanoparticles. Y. M. and A. M.: MTT assay and antiviral activity against SARS-CoV-2 using crystal violet assay. M. R. G. and A. M.: antiviral activity against SARS-CoV-2, infectivity and cell culture techniques. N. A. F.: antiviral activity against ADV-5 using PCR assay and PCR against SARS-CoV-2 under the supervision of Prof. S. A. L. M. H. E.: project reports, editing, formatting process and literature survey. F. A.: docking of potential antiviral materials into proteins of ADV-5 and SARS-CoV-2. H. A. E: maintenance Vero E6 in Lab, Cytotoxicity of tested materials on Vero E6, infectivity assay of adenovirus 5 under the supervision of Prof. S. A. L. E. A. R.: supervision. H. M. A.: supervision and reviewing manuscript. A. M.: supervision and reviewing antiviral activity against SARS-CoV-2. M. A. A.: all antiviral process against SARS-CoV-2 performed under his supervision. A. K.: supervision, reviewing, sharing in conceptualization of project and research team.
Conflicts of interest
The authors declare no competing financial interest.
Supplementary Material
Acknowledgments
This project was funded by the Egyptian Academy of Scientific Research and Technology (ASRT) under the COVID-19 Emergency Call; Ideation, Project ID 6901.
Electronic supplementary information (ESI) available: Table S1; residues interacting with ligands (CNPs, Sil, and RMV) in the predicted binding pockets of Hexon, ACE2, and Spike protein. Fig. S1; a schematic diagram of the chemical interaction of Sil@CNPs nanocomposite. See https://doi.org/10.1039/d2ra00905f
References
- Jayk Bernal A. et al., Molnupiravir for oral treatment of Covid-19 in nonhospitalized patients. N. Engl. J. Med. 2022;386(6):509–520. doi: 10.1056/NEJMoa2116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho J. et al., Antiviral activity of digoxin and ouabain against SARS-CoV-2 infection and its implication for COVID-19. Sci. Rep. 2020;10(1):1–8. doi: 10.1038/s41598-019-56847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waye M. M. Y. Sing C. W. Anti-viral drugs for human adenoviruses. Pharmaceuticals. 2010;3(10):3343–3354. doi: 10.3390/ph3103343. [DOI] [Google Scholar]
- Jindal S. Gopinath P. Nanotechnology based approaches for combatting COVID-19 viral infection. Nano Express. 2020;1(2):1–13. doi: 10.1088/2632-959X/abb714. [DOI] [Google Scholar]
- Chauhan G. et al., Nanotechnology for COVID-19: therapeutics and vaccine research. ACS Nano. 2020;14(7):7760–7782. doi: 10.1021/acsnano.0c04006. [DOI] [PubMed] [Google Scholar]
- Zhu X. et al., Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166:1–7. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauf M. A. et al., Nano-therapeutic strategies to target coronavirus. View. 2021:20200155. doi: 10.1002/VIW.20200155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla A. et al., Coronaviruses—drug discovery and therapeutic options. Nat. Rev. Drug Discovery. 2016;15(5):327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu W., et al., Acupuncture for chemotherapy-induced leukopenia: exploratory meta-analysis of randomized controlled trials, in Database of Abstracts of Reviews of Effects (DARE): Quality-assessed Reviews [Internet], Centre for Reviews and Dissemination, UK, 2007 [DOI] [PubMed] [Google Scholar]
- Peiris J. et al., Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361(9366):1319–1325. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzon M. Marsh M. Targeting viral entry as a strategy for broad-spectrum antivirals. F1000Research. 2019;8:1–10. doi: 10.12688/f1000research.19694.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. J. et al., The site of attachment in human rhinovirus 14 for antiviral agents that inhibit uncoating. Science. 1986;233(4770):1286–1293. doi: 10.1126/science.3018924. [DOI] [PubMed] [Google Scholar]
- Kobayashi K. et al., Surface engineering of nanoparticles for therapeutic applications. Polym. J. 2014;46(8):460–468. doi: 10.1038/pj.2014.40. [DOI] [Google Scholar]
- Milovanovic M., et al., Nanoparticles in antiviral therapy, in Antimicrobial nanoarchitectonics, Elsevier, 2017, pp. 383–410 [Google Scholar]
- Federico A. Dallio M. Loguercio C. Silymarin/silybin and chronic liver disease: a marriage of many years. Molecules. 2017;22(2):191. doi: 10.3390/molecules22020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.-H. et al., Antiviral activities of silymarin and derivatives. Molecules. 2019;24(8):1552. doi: 10.3390/molecules24081552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palit P. Mukhopadhyay A. Chattopadhyay D. Phyto-pharmacological perspective of Silymarin: A potential prophylactic or therapeutic agent for COVID-19, based on its promising immunomodulatory, anti-coagulant and anti-viral property. Phytother. Res. 2021:1–12. doi: 10.1002/ptr.7084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroumand H. et al., Chitosan-Based Nanoparticles Against Viral Infections. Front. Cell. Infect. Microbiol. 2021;11:175. doi: 10.3389/fcimb.2021.643953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanafy N. A. Leporatti S. El-Kemary M. A. Extraction of chlorophyll and carotenoids loaded into chitosan as potential targeted therapy and bio imaging agents for breast carcinoma. Int. J. Biol. Macromol. 2021;182:1150–1160. doi: 10.1016/j.ijbiomac.2021.03.189. [DOI] [PubMed] [Google Scholar]
- Sami El-banna F. et al., Chitosan as a natural copolymer with unique properties for the development of hydrogels. Appl. Sci. 2019;9(11):2193. doi: 10.3390/app9112193. [DOI] [Google Scholar]
- Khaliq T. et al., Self-crosslinked chitosan/κ-carrageenan-based biomimetic membranes to combat diabetic burn wound infections. Int. J. Biol. Macromol. 2022;197:157–168. doi: 10.1016/j.ijbiomac.2021.12.100. [DOI] [PubMed] [Google Scholar]
- Loutfy S. A. et al., Antiviral activity of chitosan nanoparticles encapsulating curcumin against hepatitis C virus genotype 4a in human hepatoma cell lines. Int. J. Nanomed. 2020;15:2699. doi: 10.2147/IJN.S241702. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Fernandes Queiroz M. et al., Does the use of chitosan contribute to oxalate kidney stone formation? Mar. Drugs. 2015;13(1):141–158. doi: 10.3390/md13010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-j. You J.-z. Study on the thermal decomposition of sofosbuvir. J. Anal. Appl. Pyrolysis. 2017;123:376–384. doi: 10.1016/j.jaap.2016.11.003. [DOI] [Google Scholar]
- Hasan M. et al., Preparation, characterization, and release kinetics of chitosan-coated nanoliposomes encapsulating curcumin in simulated environments. Molecules. 2019;24(10):2023. doi: 10.3390/molecules24102023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.-J. et al., Multiplexed screening of cellular uptake of gold nanoparticles using laser desorption/ionization mass spectrometry. J. Am. Chem. Soc. 2008;130(43):14139–14143. doi: 10.1021/ja805392f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmahdy E. M. et al., Molecular detection of human adenovirus in urban wastewater in Egypt and among children suffering from acute gastroenteritis. J. Water Health. 2019;17(2):287–294. doi: 10.2166/wh.2019.303. [DOI] [PubMed] [Google Scholar]
- Betancur-Galvis L. et al., Cytotoxic and antiviral activities of Colombian medicinal plant extracts of the Euphorbia genus. Mem. Inst. Oswaldo Cruz. 2002;97:541–546. doi: 10.1590/S0074-02762002000400017. [DOI] [PubMed] [Google Scholar]
- Kandeil A. et al., Coding-complete genome sequences of two sars-cov-2 isolates from egypt. Microbiol. Resour. Announce. 2020;9(22):e00489-20. doi: 10.1128/MRA.00489-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostafa A. et al., FDA-approved drugs with potent in vitro antiviral activity against severe acute respiratory syndrome coronavirus 2. Pharmaceuticals. 2020;13(12):443. doi: 10.3390/ph13120443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghaddam A. H. et al., Preventive effect of silymarin-loaded chitosan nanoparticles against global cerebral ischemia/reperfusion injury in rats. Eur. J. Pharmacol. 2020;877:173066. doi: 10.1016/j.ejphar.2020.173066. [DOI] [PubMed] [Google Scholar]
- Woo J. S. et al., Formulation and biopharmaceutical evaluation of silymarin using SMEDDS. Arch. Pharmacal Res. 2007;30(1):82–89. doi: 10.1007/BF02977782. [DOI] [PubMed] [Google Scholar]
- Calvo P. et al., Novel hydrophilic chitosan-polyethylene oxide nanoparticles as protein carriers. J. Appl. Polym. Sci. 1997;63(1):125–132. doi: 10.1002/(SICI)1097-4628(19970103)63:1<125::AID-APP13>3.0.CO;2-4. [DOI] [Google Scholar]
- Aboshanab M. H. et al., Fabrication, characterization and biological evaluation of silymarin nanoparticles against carbon tetrachloride-induced oxidative stress and genotoxicity in rats. Int. J. Pharm. 2020;587:119639. doi: 10.1016/j.ijpharm.2020.119639. [DOI] [PubMed] [Google Scholar]
- Moghaddam A. H. et al., Preventive effect of silymarin-loaded chitosan nanoparticles against global cerebral ischemia/reperfusion injury in rats. Eur. J. Pharmacol. 2020;877:173066. doi: 10.1016/j.ejphar.2020.173066. [DOI] [PubMed] [Google Scholar]
- Fernandes Queiroz M. et al., Does the use of chitosan contribute to oxalate kidney stone formation? Mar. Drugs. 2015;13(1):141–158. doi: 10.3390/md13010141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S. Singh S. K. Girotra P. Targeting silymarin for improved hepatoprotective activity through chitosan nanoparticles. Int. J. Pharm. Invest. 2014;4(4):156. doi: 10.4103/2230-973X.143113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarach N. Dodiuk H. Kenig S. Polymers in the medical antiviral front-line. Polymers. 2020;12(8):1727. doi: 10.3390/polym12081727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurunathan S. et al., Antiviral potential of nanoparticles—Can nanoparticles fight against coronaviruses? Nanomaterials. 2020;10(9):1645. doi: 10.3390/nano10091645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysenko V. et al., Nanoparticles as antiviral agents against adenoviruses. Adv. Nat. Sci.: Nanosci. Nanotechnol. 2018;9(2):025021. [Google Scholar]
- Nan Y. Wu C. Zhang Y.-J. Interplay between Janus kinase/signal transducer and activator of transcription signaling activated by type I interferons and viral antagonism. Front. Immunol. 2017;8:1758. doi: 10.3389/fimmu.2017.01758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al., Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426(6965):450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medhi R. et al., Nanoparticle-based strategies to combat COVID-19. ACS Appl. Nano Mater. 2020;3(9):8557–8580. doi: 10.1021/acsanm.0c01978. [DOI] [PubMed] [Google Scholar]
- Sharma N. Modak C. Singh P. K. Kumar R. Khatri D. Singh S. B. Underscoring the immense potential of chitosan in fighting a wide spectrum of viruses: A plausible molecule against SARS-CoV- 2? Int. J. Biol. Macromol. 2021;179:33–44. doi: 10.1016/j.ijbiomac.2021.02.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.