Abstract
Previously we reported on the HPIV2 genotype distribution in Croatia 2011–2014. Here we expand this period up to 2017 and confirm that G1a genotype has replaced G3 genotype from the period 2011–2014. Our hypothesis was that the G1a-to-G3 genotype replacement is an antibody-driven event. A cross-neutralisation with anti-HPIV2 sera specific for either G1a or G3 genotype revealed the presence of genotype-specific antigenic determinants. By the profound, in silico analyses three potential B cell epitopic regions were identified in the hemagglutinin neuraminidase (regions 314–361 and 474–490) and fusion protein (region 440–484). The region identified in the fusion protein does not show any unique site between the G1a and G3 isolates, five differentially glycosylated sites in the G1a and G3 genotype isolates were identified in epitopic regions of hemagglutinin neuraminidase. All positively selected codons were found to be located either in the region 314–316 or in the region 474–490 what indicates a strong positive selection in this region and reveals that these regions are susceptible to evolutionary pressure possibly caused by antibodies what gives a strong verification to our hypothesis that neutralising antibodies are a key determinant in the inherently complex adaptive evolution of HPIV2 in the region.
Key words: Genotype, fusion protein, hemagglutinin-neuraminidase, human parainfluenza virus type 2, neutralising antibodies
Introduction
Human parainfluenza virus type 2 (HPIV2) is a member of the genus Rubulavirus in the Paramyxoviridae family. The HPIV2 genome is a non-segmented single-stranded negative-strand RNA that contains 15 654 nucleotides. It encodes six genes for seven proteins: the nucleoprotein (NP gene), V protein/phosphoprotein (V/P gene), matrix protein (M gene), fusion protein (F gene), hemagglutinin neuraminidase (HN gene) and large protein (L gene). The major targets of the anti-HPIV2 antibodies are surface glycoproteins hemagglutinin neuraminidase and fusion protein [1, 2]. Hemagglutinin neuraminidase is anchored in the viral envelope as a tetramer (dimer of dimers) together with the fusion protein. The hemagglutinin neuraminidase has a triple role in viral infection: initial attachment to a sialic acid-containing receptor, cleaving the receptor and activating the F protein. The F protein is synthesised as a biologically inactive F0 precursor from which a prefusion metastable form of disulphide-linked F2 + F1 is formed by the proteolityc action of the cellular endoprotease (reviewed in [3]). Binding of the HN protein to the receptor initiates the F protein to undergo conformational changes which ultimately lead to the fusion of the viral and cellular membranes [4, 5] what finally leads to the intrusion of the ribonucleocapsid into the target cell.
The dynamics and evolution of HPIV2 are largely unknown. Most HPIV2 infections are treated symptomatically outside of the hospital setting. Thus the diagnosis is missing making it difficult to follow the genetic variation and evolution of the HPIV2. There are very few studies on the evolution and phylogenetic analysis of this virus [6–8]. These phylogenetic analyses identified four genotypes (G1-4). Our previous study [8] explored hospitalised HPIV2 cases in Zagreb, Croatia during 4-year period (2011–2014) and showed that the dominant genotype in this area was G3 genotype while only one strain isolated in 2014 was positioned in the distant G1a genotype. Therefore now we extended our monitoring of the HPIV2 up to 2017 in order to determine whether G3 genotype remained dominant or it was overruled by G1a genotype.
Respiratory diseases are very often the cause of hospitalisations worldwide. The major cause is infection with a respiratory syncytial virus (RSV), while human parainfluenza viruses (HPIVs) infections are the second major cause of hospitalisation for respiratory tract illness in young children [9]. HPIV are spread by respiratory secretions from infected persons or contact with contaminated surfaces. Infections with these viruses are associated with significant morbidity and at least as much acute respiratory illness (ARI) in infants and young children as [10–13]. HPIV2 causes ARI (croup, pneumonia, bronchitis and bronchiolitis) in infants, young children and immunocompromised elderlies [14] although with lower prevalence rate than HPIV3 and HPIV1. Infections with HPIV2 are also recognised less often because of its mild nature, especially, in otherwise healthy adults. For that reason, in Croatia, the diagnosis for HPIV2 is conducted only for hospitalised children or adults with immunocompromised conditions. Most HPIV2 infection have been detected in autumn or early winter at 1 or 2-year interval. The overall relative contribution to the burden of disease in infants and young children is estimated at approx. 7:3.5:2:1 for HPIV3, HPIV1, HPIV2 and HPIV4, respectively [10, 15–17]. Also, these viruses are proven to be associated with nosocomial ARI in immunocompromised patients [16, 18, 19]. In spite of the impact of these viruses on the public health no licensed vaccine or any other specific anti-viral treatment against any type of HPIV exists because of the incomplete knowledge of the immune response against these viruses. Almost all children encounter HPIVs within the first few years after birth, but immunity seems to be incomplete and re-infections occur throughout life. However, after primary infection, a certain level of protection develops which is able to prevent severe disease later in life but it is unable to completely suppress the infection [9]. The reason for this incomplete protection may be a genetic drift of the virus as we imply in this study.
In this study, we examine the role of neutralising antibodies as the promoter of the inter-seasonal dynamics of the HPIV2 and potentially as s primary cause of the incomplete immunity to re-infections with viruses of distant genotypes.
Materials and methods
Isolation of virus from clinical specimens, amplification of the F and HN genes and sequencing
Clinical specimens were obtained from 38 patients hospitalised for respiratory infections. Samples were collected during 2011–2017 as nasopharyngeal secretions (details in Table 1) and detected as HPIV2 positive by direct fluorescence assays (DFA Light Diagnostics, Chemicon International, Temecula, CA, USA). None of the samples was found to be a co-infection with another respiratory pathogen. This study was approved by the ethics committee of the University Hospital for Infectious Diseases ‘Dr Fran Mihaljević’.
Table 1.
HPIV2 isolates collected from patients during 2011–2017 in Zagreb, Croatia
| Isolate namea | Ageb | Collection date | Diagnosisc | GenBank Accessiond |
|---|---|---|---|---|
| HPIV2i/Zagreb.HR/47.11(16406) | 8 m | Oct 2011 | C | KT315655 |
| HPIV2i/Zagreb.HR/47.11(16475) | 1 y 11 m | Oct 2011 | P | KT315656 |
| HPIV2i/Zagreb.HR/49.11(16967) | 7 m | Nov 2011 | C | KT315657 |
| HPIV2i/Zagreb.HR/49.11(16971) | 1 y | Nov 2011 | URI | KT315658 |
| HPIV2i/Zagreb.HR/49.11(17034) | 7 m | Nov 2011 | URI | KT315659 |
| HPIV2i/Zagreb.HR/28.12(2706) | 1 y 9 m | Jun 2012 | C | KT315660 |
| HPIV2i/Zagreb.HR/29.12(2725) | 2 m | Jul 2012 | URI | KT595232 |
| HPIV2i/Zagreb.HR/38.12(2858) | 6 m | Sep 2012 | B | KT315661 |
| HPIV2i/Zagreb.HR/41.12(2950) | 5 m | Sep 2012 | C | KT315662 |
| HPIV2i/Zagreb.HR/42.12(2995) | 7 y | Oct 2012 | URI | KT595233 |
| HPIV2i/Zagreb.HR/44.12(3036) | 4 y 4 m | Oct 2012 | C | KT595234 |
| HPIV2i/Zagreb.HR/50.12(3257) | 8 m | Nov 2012 | B | KT315663 |
| HPIV2i/Zagreb.HR/50.12(3263) | 8 m | Nov 2012 | URI | KT315664 |
| HPIV2i/Zagreb.HR/51.12(3316) | 2 m | Dec 2012 | URI | KT315665 |
| HPIV2i/Zagreb.HR/42.14(1472) | 6 m | Oct 2014 | URI | KT315666 |
| HPIV2i/Zagreb.HR/47.14(1688) | 5 y 6 m | Nov 2014 | P | KT315667 |
| HPIV2i/Zagreb.HR/47.15(1779) | 5 y 7 m | Nov 2015 | P | MF163150 |
| HPIV2i/Zagreb.HR/49.15(1849) | 2 y 3 m | Dec 2015 | URI | MF163151 |
| HPIV2i/Zagreb.HR/03.16(222) | 2 y 6 m | Jan 2016 | P | MG460766 |
| HPIV2i/Zagreb.HR/28.16(1623) | 1 y 10 m | Jul 2016 | C | MG460767 |
| HPIV2i/Zagreb.HR/37.16(1827) | 4 y | Sep 2016 | URI | MG460768 |
| HPIV2i/Zagreb.HR/40.16(1916) | 3 y 4 m | Oct 2016 | URI | MG460769 |
| HPIV2i/Zagreb.HR/41.16(1939) | 3 y 8 m | Oct 2016 | C | MG460770 |
| HPIV2i/Zagreb.HR/42.16(1995) | 1 y 1 m | Oct 2016 | C | MG460771 |
| HPIV2i/Zagreb.HR/44.16(2085) | 2 y 6 m | Nov 2016 | URI | MG460772 |
| HPIV2i/Zagreb.HR/44.16(2115) | 1 y 2 m | Nov 2016 | C | MG460773 |
| HPIV2i/Zagreb.HR/45.16(2165) | 15 y 9 m | Nov 2016 | URI | MG460774 |
| HPIV2i/Zagreb.HR/46.16(2188) | 9 m | Nov 2016 | C | MG460775 |
| HPIV2i/Zagreb.HR/46.16(2209) | 1 y 4 m | Nov 2016 | C | MG460776 |
| HPIV2i/Zagreb.HR/46.16(2241) | 57 y 4 m | Nov 2016 | URI | MG460777 |
| HPIV2i/Zagreb.HR/49.16(2400) | 17 y 3 m | Dec 2016 | URI | MG460778 |
| HPIV2i/Zagreb.HR/50.16(2469) | 2 m | Dec 2016 | P | MG460779 |
| HPIV2i/Zagreb.HR/50.16(2513) | 1 y 4 m | Dec 2016 | URI | MG460780 |
| HPIV2i/Zagreb.HR/50.16(2514) | 9 m | Dec 2016 | URI | MG460781 |
| HPIV2i/Zagreb.HR/50.16(2534) | 4 m | Dec 2016 | B | MG460782 |
| HPIV2i/Zagreb.HR/03.17(208) | 1 y | Jan 2017 | B | MG460783 |
| HPIV2i/Zagreb.HR/03.17(228) | 2 y 10 m | Jan 2017 | URI | MG460784 |
| HPIV2i/Zagreb.HR/07.17(705) | 5 m | Feb 2017 | URI | MG460785 |
Isolate number is given in parentheses.
m – month; y – year.
P-pneumonia, B-bronchiolitis, C-croup, URI-upper respiratory tract infection.
Accession number of the sequence of the F and HN genes submitted to the GenBank.
The procedure to obtain the sequences of the F and HN genes consisted of the virus isolation from each specimen on Vero cells in the presence of trypsin in order to decrease the number of mutations during cultivation in the cell culture as shown in [8]. A number of isolated viruses vs. obtained clinical specimen was: 5/5 (2011), 9/11 (2012), 2/6 (2014), 2/4 (2015), 17/18 (2016) and 3/3 (2017). Isolation of viral RNA, amplification of the F and HN genes and Sanger sequencing of amplified segments. All procedures have been previously described in [8].
Phylogenetic analyses of the F and HN genes
Accession numbers of sequences obtained from the GenBank which were included in analysis are DQ072586-DQ 072589, JF912194-JF912196, NC_003443, AF533011, D00865, AF533012, AB176531, AB189948-AB189953, KT315655-KT315667 and KT595232-KT595234.
Alignments were performed by CloneManager Suite software (Scientific&Educational Software, USA).
For phylogenetic analyses, the coding region of the HN gene sequence was used. Multiple sequence alignments were made using the CLSUTAL W program integrated in software MEGA6.0 (http://www.megasoftware.net). The best evolutionary model for phylogenetic inference was determined by using Model selection implemented in MEGA6.0. The phylogenetic tree was generated in MEGA6.0 using maximum likelihood (ML) analysis with the Kimura 2-parametar substitution model and 1000 bootstrap replicates.
Immunisation of guinea pigs with HPIV2
Two strains of HPIV2 were used for immunisation of guinea pigs: HPIV2i/Zagreb.HR/42.14(1472) (acc.nr. KT315666) and HPIV2i/Zagreb.HR/47.14(1688) (acc.nr. KT315667). HPIV2i/Zagreb.HR/42.14(1472) has been previously shown to belong to genotype G3 and HPIV2i/Zagreb.HR/47.14(1688) belongs to genotype G1a [8].
All animal work was in accordance to Croatian Low on Animal Welfare (2017) which strictly complies with EC Directive (2010/63/EU). Female guinea pigs weighing 300–400 g (Institute of Immunology Inc., Zagreb, Croatia) were housed in a 12-h light/12-h dark cycle and a constant temperature of 22 °C. A standard guinea pig diets (Muchedola srl, Milano, Italy) and water were supplied ad libitum during all immunisation period.
Antigen for immunisation was prepared by growing the two HPIV2 strains on Vero cells, centrifugation of the cell culture supernatants at low speed for 10 min and then by ultracentrifugation at 28 000 rpm in SW28 rotor (Beckman Coulter, Indianapolis, USA) for 2 h. Finally, the pellet was resuspended in PBS, pH 7.4. Six animals were immunised with each HPIV2 strain while the mock group consisted of three animals received PBS only. Animals were given three s.c. immunisations (days 0, 18 and 36) each containing 4.3 logPFU. Fifty days following prime immunisation guinea pigs’ blood was collected by heart puncture and sera were prepared for further serological analyses by centrifugation at 1200g at 4 °C for 30 min and decomplementation by heating at 56 °C for 30 min.
Plaque reduction neutralisation test (PRNT)
PRNT was performed as in [20], with some modifications. Test sera were prepared in six fourfold dilutions starting from at least one dilution completely neutralising added virus and ending with at least one dilution having no neutralising capacity. The virus concentration was adjusted to 35 ± 10 PFU/0.1 ml with MEM-H with 2.5% FCS. The test was performed in 24-well plates in which Vero cells were seeded at concentration 3.5 × 105 cells/well 24 h prior to the addition of mixture serum/virus. Each dilution of the serum was mixed with an equal volume of virus and incubated at 35 °C for 1 h. Then the medium was removed from the plate and 0.2 ml of the mixture was transferred in four wells in the plate. Each assay run included virus as a positive control and medium used for diluting the sera and viruses as a negative control. The plates were incubated at 35 °C for 1.5 h, the mixture serum/virus was removed and the cells were washed twice with medium without serum. Finally, cells were overlayed with 0.75 ml of a semisolid medium consisting of 1 v/v 2 × MEM-H without phenol red, 1 v/v 1.5% Noble agar (Sigma) and 0.75 µg/ml acetylated trypsin (Sigma). Plates were incubated at 35 °C in a humidified atmosphere of 5% CO2. After six days the monolayers were stained with 0.5 ml of 0.05% neutral red (Sigma) and plaques were counted. Fifty percent end point titres were calculated using the Kärber formula: log10 ND50 = −max−Δ(∑p−0.5), where max is the log10 of the highest dilution of serum in the test, Δ is log10 of the dilution factor (for fourfold dilutions is −0.6) and ∑p is total number of plaques divided by the number of plaques in positive control sample.
A two-tailed unpaired Students’ t test was used to calculate statistical significance.
In silico B cell epitope prediction
Protein structures of the fusion protein and hemagglutinin neuraminidase of HPIV2i/Zagreb.HR/42.14(1472) and HPIV2i/Zagreb.HR/47.14(1688) was modelled using a homology SWISS-model platform [21–23] (http://swissmodel.expasy.org) in order to create PDB structures.
Three different servers were used to predict conformational epitopes in the fusion protein and hemagglutinin neuraminidase: (a) DiscoTope 2.0 server [24], (b) ElliPro [25] and (c) BepiPred-2.0 [26]. DiscoTope 2.0 and ElliPro servers were used with default settings and BepiPred-2.0 was used at treshold 0.55.
Only epitopic regions which were predicted by all three servers were selected.
Analysis of glycosylation sites within the fusion protein and hemagglutinin neuraminidase
Complete fusion protein and hemagglutinin neuraminidase sequence were submitted for prediction of N-linked and O-linked glycosylation sites by using glycosylation predictor program which identifies pairwise patterns surrounding glycosylation sites and use an odds ratio to weight their propensity of association with modified residues [27] (http://comp.chem.nottingham.ac.uk/glyco/).
Natural selection analysis
To evaluate the selection pressure effect on each codon/amino site in the predicted epitopic regions of the hemagglutinin neuraminidase the HN ORFs of all strains involved in the phylogenetic analyses were analysed by computing ratio between non-synonymous and synonymous substitutions. The selection analyses consisted of four independent maximum likelihood methods from the Datamonkey server [28] (http://www.datamonkey.org/): a single-likelihood ancestor counting (SLAC), a fixed-effects likelihood (FEL), an internal branch fixed-effects likelihood (IFEL) and a random effects likelihood (REL), which estimate synonymous and nonsynonymous substitution rates at every codon. Sites were considered positively selected if they met the cut-off criteria of P-value of <0.1 for SLAC, FEL and IFEL, or Bayes factors >50 for REL.
Results
Distribution of HPIV2 genotypes in Croatia in the period 2011–2017
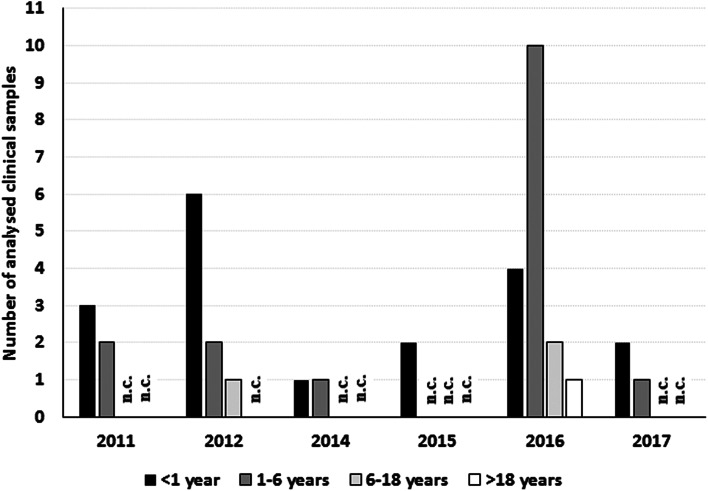
A total of 38 HPIV2 specimens were collected from hospitalised patients in Zagreb, Croatia, in period 2011–2017: 16 isolates from period 2011 to 2014 were previously described in [8] and the new 22 isolates have been collected and isolated in the period 2015–2017. The age distribution for each season (Fig. 1) shows that age groups were evenly distributed throughout the seasons with the exception of the season 2016 where the number of patients in the age group 1–6 years is larger than in the age group <1 year. Unfortunately, the data about possible reinfection of patients from the age group 1–6 years are unavailable so it was not possible to further explain if these patients had a primary infection or they were re-infected.
Fig. 1.
Age distribution of the patients from whom the samples were collected. n.c., none collected.
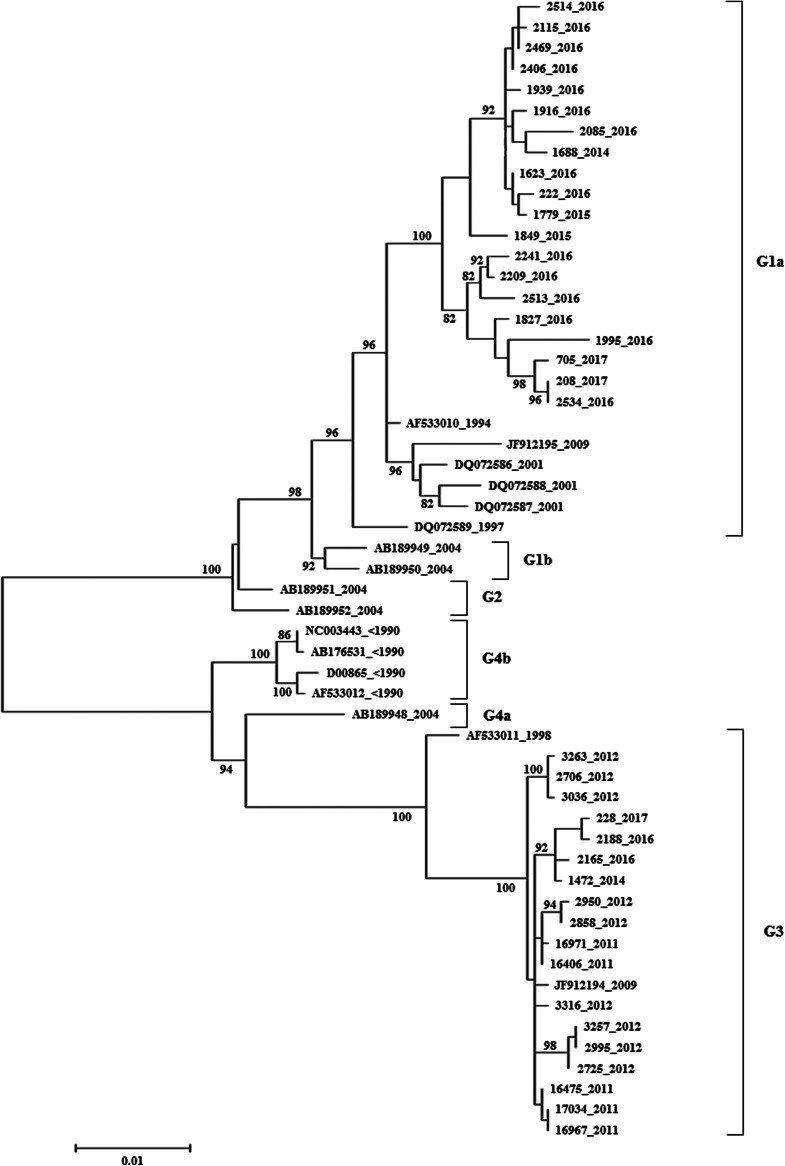
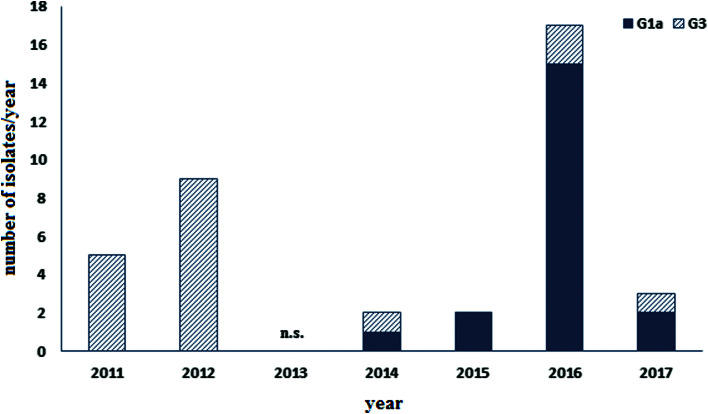
Our previous study [8] showed that in 2014 one isolate of genotype G1a appeared while the other isolate from 2014 and earlier isolates belong to genotype G3. This urged further analysis in order to confirm the replacement of the G3 genotype with G1a genotype in this area. The phylogenetic analysis for all 38 samples was performed and the phylogenetic tree is presented in Figure 2. The analysis revealed that the vast majority of isolates collected after 2014 are placed in genotype G1a (19/22, Figs 2 and 3). However, the G3 genotype has been still sporadically circulating in the population but with much lower intensity (3/22, Figs 2 and 3) indicating that G1a genotype is been replacing G3 genotype.
Fig. 2.
Maximum likelihood phylogenetic tree of the HN gene ORF of HPIV2 strains isolated in Croatia 2011–2017 and sequences accessible in the GenBank constructed by the MEGA6.0 with 1000 bootstrap replicates. Croatian isolates are presented with their isolate numbers and sequences retrieved from the GenBank are presented with their accession numbers. The year of isolation is indicated for each sequence. The genotypes are indicated by the brackets on the right side. Only bootstrap values over 80% are displayed at the branch nodes. The scale bar indicates 0.01 nt differences per site, over the indicated region.
Fig. 3.
Number of isolates from Croatia 2011–2017 belonging to G1a or G3 genotype. n.s., no specimen obtained.
Genotype specific neutralisation potential of guinea pigs’ immunised with different HPIV2 strains
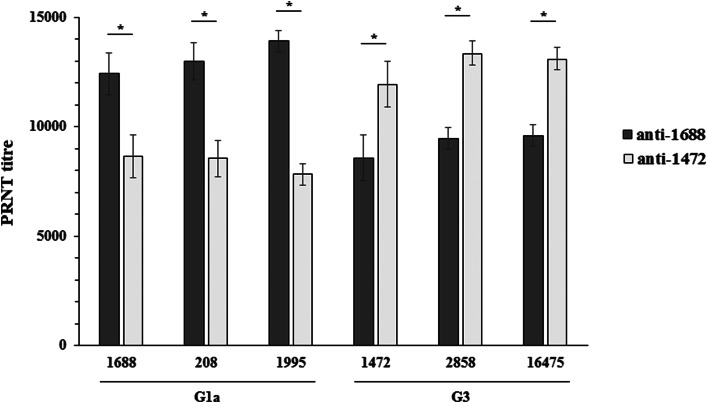
A possible reason of G1a genotype becoming dominant over the G3 genotype is an incomplete antigenic match between the two genotypes. To analyse this, an immunisation of guinea pigs with two HPIV2 strains was performed in order to obtain neutralising sera specific for different genotypes. Both viruses used for immunisation were isolated from a specimen collected in 2014 but belonged to different genotype: HPIV2i/Zagreb.HR/42.14(1472) (G3 genotype) and HPIV2i/Zagreb.HR/47.14(1688) (G1a genotype). The results of the PRNT with guinea pigs’ sera and the two viruses are shown in Figure 4. Sera of animals immunised with either virus had strong neutralising activity. However, sera specific for the genotype G3 1472 virus had significantly lower neutralisation capacity for the genotype G1a 1688 virus (PRTN50 titres 11 935.5 ± 1 046.5 for 1472 virus vs. 8 654.7 ± 972.1 for 1688 virus, P < 0.01, Fig. 4) and vice versa, sera specific for the genotype G1a had significantly lower capacity to neutralise virus of the genotype G3 (PRTN50 titres 8566.1 ± 925.3 for 1472 virus vs. 12 407.7 ± 1389.7 for 1688 virus, P < 0.01, Fig. 4). To confirm that this finding is not specific for these two viruses only but can be applied at the genotype level, two more viruses from both, G1a and G3, genotypes were neutralised with these sera. Figure 4 shows that the neutralisation capacity of the sera is comparable as long as the genotype of the virus used for immunisation matches the virus used in neutralisation test.
Fig. 4.
Neutralisation of HPIV2i/Zagreb.HR/47.14(1688), HPIV2i/Zagreb.HR/03.17(208), HPIV2i/Zagreb.HR/42.16(1995), HPIV2i/Zagreb.HR/42.14(1472), HPIV2i/Zagreb.HR/38.12(2858) and HPIV2i/Zagreb.HR/47.11(16 475) with guinea pigs’ sera immunised with either HPIV2i/Zagreb.HR/47.14(1688) or HPIV2i/Zagreb.HR/42.14(1472) (n = 6 for each virus). Values are mean ± STD. *P < 0.001.
These findings indicate that there are some antigenic differences between the two genotypes which are the cause of the lower neutralisation specificity and possibly the cause of waning of the genotype G3 from the population and establishing the dominance of the genotype G1a.
Analysis of in silico predicted conformational B cell epitopes in the hemagglutinin neuraminidase and fusion protein
Although the attachment protein is the major target of neutralising antibodies, antibodies raised against F proteins are also able to neutralise infections [1, 2]. To further analyse the mechanism of how one HPIV2 genotype is replacing another, conformational B cell epitopes for two viral surface glycoproteins, fusion protein and hemagglutinin neuraminidase, were predicted in silico. To increase the relevance of the predicted epitopic regions three servers which predict conformational B cell epitopes were used: DiscoTope2.0, ElliPro and BepiPred-2.0. Only those regions predicted to contain epitopes which were predicted by all three tools were selected.
Analysis of the hemagglutinin neuraminidase
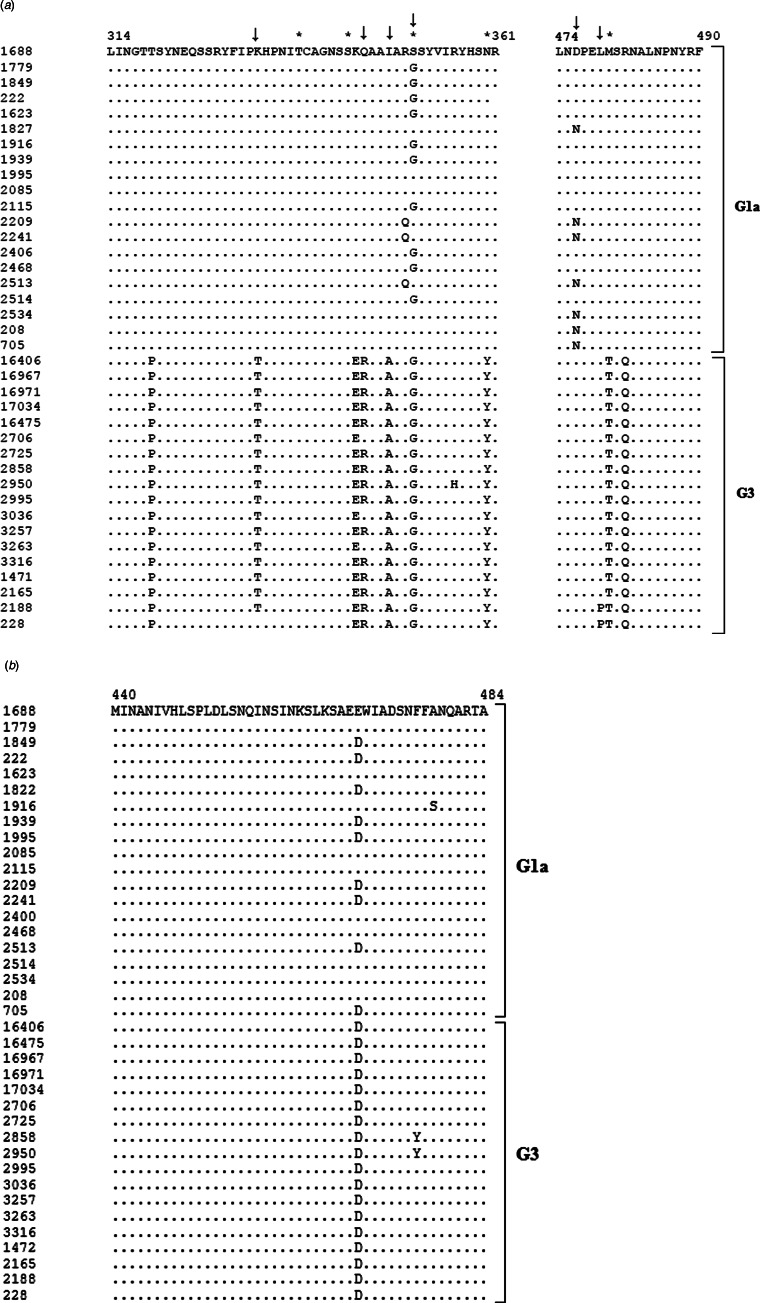
The putative hemagglutinin neuraminidase sequences were subjected to homology modelling what yielded a homo-tetramer model with >48% of homology. A PDB structure was submitted to in silico prediction of potential epitopes. Each search resulted in several predicted epitopes of different size but only two regions were identified as potential epitopes by all three servers: a region between amino acids 314 and 361 and a region between amino acids 474 and 490 (Table 2). Although there are 24 unique sites throughout the hemagglutinin neuraminidase (24/571 amino acid residues (Supplementary Fig. S1)) alignments of the predicted epitope sequences show that these two regions contain amino acid residues unique for the genotype (Fig. 5a). Additionally, these two potential epitopic regions were searched for N-linked and O-linked glycosylation sites. Five sites with different glycosylation status for viruses 1472 and 1688 were identified: 338Thr, 244Ser and 481Thr were glycosylated in 1688 and 351Ser and 360Asn were glycosylated in 1472 (Table 3). Three of the differentially glycosylated sites are the genotype unique amino acid residues (Ser351Gly, Asn360Tyr, Met481Thr), while the other two differentially glycosylated sites are not but might be influenced by the neighbouring unique residues (Table 3).
Table 2.
In silico predicted B cell epitopes for hemagglutinin neuraminidase and fusion protein.
| Protein | Epitope region (aa) | Server | Epitope sequence (1472 vs.1688) |
|---|---|---|---|
| Hemagglutinin neuraminidase | 314–361 | DiscoTope2.0 | 317-GTPSYNE-323 |
| 317-GTTSYNE-323 | |||
| BepiPred-2.0 | 317-GTPSYNEQSSRYFIP-331 | ||
| 317-GTTSYNEQSSRYFIP-331 | |||
| ElliPro | 314-LINGTPSYNEQSSRYFIPTHPNITCAGNSSERAAAARGSYVIRYHSYR-361 | ||
| 314-LINGTTSYNEQSSRYFIPKHPNITCAGNSSKQAAIARSSYVIRYHSNR-361 | |||
| 474–490 | DiscoTope2.0 | 481-SQN-483 | |
| 481-SRN-483 | |||
| BepiPred-2.0 | 477-PELTSQNALN-486 | ||
| 477-PELMSRNALN-486 | |||
| ElliPro | 474-LNDPELTSQNALNPNYR-490 | ||
| 474-LNDPELMSRNALNPNYR-490 | |||
| Fusion protein | 440–484 | DiscoTope2.0 | 450-PLDLSNQINSINKSLKSAEDWIADSNFFANQA-481 |
| 450-PLDLSNQINSINKSLKSAEEWIADSNFFANQA-481 | |||
| BepiPred-2.0 | 440-MINANIVHLSPLDLSNQINSINKSLKSAEDWIADSNFFANQARTA-484 | ||
| 440-MINANIVHLSPLDLSNQINSINKSLKSAEEWIADSNFFANQARTA-484 | |||
| ElliPro | 446-VHLSPLDLSNQINSINKSLKSAEDWIADSNFFANQA-481 | ||
| 446-VHLSPLDLSNQINSINKSLKSAEEWIADSNFFANQA-481 |
Different residues in HPIV2 1472 and HPIV2 1688 are bold and underlined.
Fig. 5.
Predicted conformational epitopes of (a) hemagglutinin neuraminidase and (b) fusion protein of Croatian isolates belonging to genotypes G1a and G3. The genotypes are indicated by the brackets on the right side. Non-matching residues are shown. Potentially glycosylated residues are marked with asterisk (*) and positively selected codons are marked with an arrow (↓).
Table 3.
Glycosylated and positively selected sites in predicted epitopes in hemagglutinin neuraminidase
| Epitope sequence (314–361) | Glycosylated sitea | Positively selected site | Epitope sequence (474–490) | Glycosylated sitea | Positively selected site | ||
|---|---|---|---|---|---|---|---|
| 1472 | 1688 | 1472 | 1688 | ||||
| L | L | − | − | L | L | − | − |
| I | I | − | − | N | N | − | − |
| N | N | − | − | D | D | − | + |
| G | G | − | − | P | P | − | − |
| T | T | − | − | E | E | − | − |
| P | T | − | − | L | L | − | + |
| S | S | − | − | T | M | +(1688) | − |
| Y | Y | − | − | S | S | − | − |
| N | N | − | − | Q | R | − | − |
| E | E | − | − | N | N | − | − |
| Q | Q | − | − | A | A | − | − |
| S | S | − | − | L | L | − | − |
| S | S | − | − | N | N | − | − |
| R | R | − | − | P | P | − | − |
| Y | Y | − | − | N | N | − | − |
| F | F | − | − | Y | Y | − | − |
| I | I | − | − | R | R | − | − |
| P | P | − | − | ||||
| T | K | − | + | ||||
| H | H | − | − | ||||
| P | P | − | − | ||||
| N | N | − | − | ||||
| I | I | − | − | ||||
| T | T | +(1688) | − | ||||
| C | C | − | − | ||||
| A | A | − | − | ||||
| G | G | − | − | ||||
| N | N | − | − | ||||
| S | S | − | − | ||||
| S | S | +(1688) | − | ||||
| E | K | − | − | ||||
| R | Q | − | + | ||||
| A | A | − | − | ||||
| A | A | − | − | ||||
| A | I | − | + | ||||
| A | A | − | − | ||||
| R | R | − | − | ||||
| G | S | +(1472) | + | ||||
| S | S | − | − | ||||
| Y | Y | − | − | ||||
| V | V | − | − | ||||
| I | I | − | − | ||||
| R | R | − | − | ||||
| Y | Y | − | − | ||||
| H | H | − | − | ||||
| S | S | − | − | ||||
| Y | N | +(1688) | − | ||||
| R | R | − | − | ||||
Virus with glycosylated site is in parenthesis.
Positive selection analysis of epitopic sites is advantageous because the mutations in these sites are highly favourable for adaptive evolution. To identify the existence of positive selection pressure at individual codon sites, four likelihood procedures (SLAC, FEL, IFEL, REL) were used for the analysis of the in silico identified epitope regions 314–361 and 474–490. The analysis was performed with the complete HN gene sequences of all HPIV2 used for phylogenetic analysis in section Distribution of HPIV2 genotypes in Croatia in the period 2011–2017. SLAC and FEL were not able to identify any positively selected site. In contrast, IFEL and REL identified six positively selected codons (Table 3, Fig. 5a), four of which encoded for amino acids located in the epitope region 314–361 and differ between HPIV2 1472 and HPIV2 1688 (Thr333Lys, Arg346Gln, Ala349Ile, Gly351Ser). The other two positively selected codons encoded for amino acid residues in the epitope region 474–490, but are identical in both HPIV2 1472 and HPIV2 1688.
Analysis of the fusion protein
The homology modelling of the putative fusion protein sequence resulted in a homo-tetramer model with >51% of homology. A PDB structure was submitted to in silico prediction of potential epitopes. A search of potential epitopes in the fusion protein resulted in a single region between amino acids 440 and 484 which were identified by all three epitope predictor servers (Fig. 5b, Table 2). The fusion protein is more conserved than hemagglutinin neuraminidase and there are only 11 unique sites throughout the fusion protein (11/551 amino acid residues (Supplementary Fig. S2)). None of them is located in the predicted epitopic region 440–484 although there is a difference Asp469Glu between HPIV2 1472 and HPIV2 1688 (Table 2) but this substitution is also present in several G1a isolates (Fig. 5b). This region was subjected to inspection for potential glycosylation sites but no difference was found for the two viruses.
Since it seems that the difference in the neutralisation capacity between sera of guinea pigs immunised with the two HPIV2 had no origin in the fusion protein, evolutional pressure selection was not analysed for this gene/protein.
Discussion
Human parainfluenza viruses are important causes of respiratory tract diseases affecting both upper and lower respiratory tract. HPIV2 infection sometimes necessitates hospitalisations, especially in children under 2 years of age or immunocompromised patients and therefore cannot be marginalised. Nevertheless, there are very few reports on the epidemiology and evolutionary dynamics of this virus. Phylogenetic analysis [7] identified two clusters within HPIV2 isolates but the low number of isolates gave poorly informative phylogenetic tree. Later, phylogenetic tree with more different worldwide HPIV2 isolates was constructed and suggested the existence of four clusters (G1–4) [6]. We have added 38 new HPIV2 isolates ([8] and this study) and thus have generated adequate number of HPIV2 sequences to create a solid phylogenetic framework (Fig. 2) which confirms the distribution of HPIV2 isolates into four genotypes identified by [6]. Our previous study of genetic diversity of HPIV2 in Croatia between 2011 and 2014 betoken that the G1a genotype would take over the dominance of the G3 genotype in this period [8]. This has been proven in the study presented here which expanded the period of analysis up to 2017. Although the G1a genotype is now dominant, the G3 genotype can still be sporadically found among the isolates (Fig. 3). A limited number of HPIV2 isolates in this study could be misleading. However, the number of collected isolates was relatively high in 3 years: 5 isolates in 2011, 9 isolates in 2012 and 17 isolates in 2016, what reassures that conclusions drawn out from this study are relevant. Although we analysed only hospitalised cases, they are a projection of the composition of the viruses circulating in the population. Furthermore, the severity of the disease demanding hospitalisation of otherwise healthy children indicates that there could be a shift in the virus population. The overall age distribution in each season was uniform except in 2016. The high number of patients from 1 to 6 years of age in this season can be explained as either coincidentally increased number of patients which later then usually encountered HPIV2 for the first time or as secondary infection which caused a severe disease due to the shift from G3 genotype to G1a genotype which resulted in an incomplete matching of antibodies developed during primary infection (G3) and the antigen causing the disease in the secondary infection (G1a). From one of the patients in 2016 an HPIV2 belonging to genotype G3 was isolated. This patient was hospitalised at the age of almost 16. In this case the G3 infection might have been a secondary infection while primary infection could have occurred more than a decade ago, at the time when unknown HPIV2 genotype was circulating in the region. Although the number of samples is relatively low, we observed two seasons with increased number of HPIV2 infections, 2012 and 2016. Such seasonal waves may occur at the time when a new genotype enters the population, wedges out the genotype which dominated until then and cause more severe disease due to incomplete antigenic matching.
While the evolutionary processes shaping HPIV2 virus diversity within one season (autumn and early winter) may be a stochastic process, here our hypothesis was that the replacement of one genotype with another across epidemic seasons is an immune-driven process with specific neutralising antibodies as a key determinant. Our hypothesis was based on the finding that there is an antigenic diversity of the hemagglutinin neuraminidase of different HPIV2 isolates [29, 30]. Also, [31] found that monoclonal antibodies which effectively neutralised the activity of hemagglutinin neuraminidase of the prototype Greer strain isolated in 1955 failed to show any detectable binding or neutralising activity against a number of natural isolates collected from 1986 and 1987. To test our hypothesis, guinea pigs were immunised with HPIV2 isolates belonging to either G1a or G3 genotype. Although epitopes recognised by sera of different species can differ, it was unfeasible to obtain undisputed human anti-HPIV2 sera specific for a single genotype since many HPIV2 infections are treated as a common cold and it is impossible to distinguish a pre-existing antibodies from those of acute infection. Also, we analysed mainly the effect of IgG present in sera while IgA may have a more important role since mucosal surfaces are primary sites of HPIV2 infection. A cross-neutralisation with guinea pigs’ anti-HPIV2 sera revealed the presence of genotype-specific antigenic determinants and confirmed our hypothesis. Similarly, it was suggested previously that vaccine raised specific antibodies in the population may lead to the replacement of the endemic genotypes with new genotype variants for Japanese encephalitis virus and measles virus [32–34].
To analyse in depths the mechanisms of differential recognition of HPIV2 by different sera, potential B cell epitopes were predicted in silico. Since attachment protein hemagglutinin neuraminidase is the major target of neutralising antibodies and antibodies to fusion protein can have neutralising activity [2] these two proteins were analysed in depth. Hemagglutinin neuraminidase is assembled as a tetramer and fusion protein as trimer on the surface of the viral particle. Such oligomeric states form quaternary structures which shape specific antigenic sites often not recognised in the monomeric form. As a result, antiviral antibodies against the extracellular domain of surface proteins predominantly recognise conformational epitopes [1]. To increase the relevance of predicted epitopes, three different methods for prediction of conformational B cell epitopes were used and only those epitopes which were predicted by all three methods were selected for further analysis. Two and one epitopic regions were identified for hemagglutinin neuraminidase and fusion protein, respectively. To analyse the origin of the antigenic difference between the two HPIV2 genotypes as the cause of the genotype replacement, a glycosylation and positive selection of these regions were analysed in more detail. Profound analyses of the predicted epitopes in the hemagglutinin neuraminidase indicate that indeed these regions (especially epitopic region 314–361) might be responsible for the decreased match between G1a and G3 genotypes. Five differentially glycosylated sites were found: four and one for epitopic regions 314–361 and 474–490, respectively. Differential glycosylation may be associated with feeble recognition of specific epitopes by changing or shielding the contact surface between specific antibody and antigen. A strong positive selection in this region (all positively selected codons identified in the hemagglutinin neuraminidase are located either in the region 314–316 or in the region 474–490) reveals that these regions are susceptible to evolutionary pressure possibly caused by antibodies what gives a strong verification to our hypothesis that neutralising antibodies are a key determinant in the inherently complex adaptive evolution of HPIV2 in the region. Hemagglutinin neuraminidase and fusion protein are major targets of neutralisation antibodies which give a long-term protective immunity against human and animal paramyxoviruses [35–39]. Although fusion protein can be a target of selection process [40] here it seems that fusion protein has a minor role in the selection process since a unique residue for the epitopic region of this protein was not identified. This finding additionally indicates that hemagglutin neuraminidase is the main target of neutralising antibodies while fusion protein has probably a very minor role in this process in HPIV2 infection.
Although the work presented here identified two epitopic regions in hemagglutinin neuraminidase and one in fusion protein there are most certainly more epitopes in these two proteins which are recognised by the neutralising antibodies and which may differ between viruses of different genotypes. There are numerous genotype unique residues in hemagglutinin neuraminidase (Supplementary Fig. S1) which may cause the viruses of the two genotypes to further diverge from each other.
The current progress in the development of human HPIV vaccine is reviewed by [41]. It is not very likely that vaccine against HPIV2 will be developed as a monovalent vaccine, rather it would be combined with another virus or viruses like HPIV1 or 3, RSV or similar. Whatever the approach (attenuated vaccine, T-cell inducing vaccines, vaccines targeting less variable proteins like nucleoprotein or matrix protein etc.), an extreme care should be taken considering the antigenic composition of the vaccine, the measures how to control it and readapt according to continuous virus surveillance of the virus circulating in the population.
It will be of interest to monitor the circulation of HPIV2 in the population to see if these two genotypes are exchanging each other or a new, third genotype will replace the current dominance of G1a.
Acknowledgements
This work has been supported by the Croatian Science Foundation under the project number 6255 and by the grant ‘Strengthening the capacity of CerVirVac for research in virus immunology and vaccinology’, KK.01.1.1.01.0006, awarded to the Scientific Centre of Excellence for Virus Immunology and Vaccines and co-financed by the European Regional Development Fund.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268818001693.
click here to view supplementary material
Conflict of interest
Authors declare no conflict of interest.
References
- 1.Iwasaki A and Medzihitov R (2013) Innate response to viral infections. In Knipe DM and Howley PM (eds), Fields Virology, 6th Edn. Philadelphia: Wolters Kluwer, pp. 189–253. [Google Scholar]
- 2.Graham BS, Crowe JE and Ledgerwood JE (2013) Immunization against viral diseases. In Knipe DM and Howley PM (eds), Fields Virology, 6th Edn. Philadelphia: Wolters Kluwer, pp. 374–413. [Google Scholar]
- 3.Chang A and Dutch RE (2012) Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4, 613–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamb RA, Paterson RG and Jardetzky TS (2006) Paramyxovirus membrane fusion: lessons from the F and HN atomic structures. Virology 344, 30–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bose S et al. (2013) Mutations in the parainfluenza virus 5 fusion protein reveal domains important for fusion triggering and metastability. Journal of Virology 87, 13520–13531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Almajhdi FN, Alshaman MS and Amer HM (2012) Human parainfluenza virus type 2 hemagglutinin-neuramindase gene: sequence and phylogenetic analysis of the Saudi strain Riyadh 105/2009. Virology Journal 9, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Terrier O et al. (2008) Characterization of naturally occurring parainfluenza virus type 2 (hPIV-2) variants. Journal of Clinical Virology 43, 86–92. [DOI] [PubMed] [Google Scholar]
- 8.Šantak M et al. (2016) Genetic diversity among human parainfluenza virus type 2 isolated in Croatia between 2011 and 2014. Journal of Medical Virology 88, 1733–1741. [DOI] [PubMed] [Google Scholar]
- 9.Hall CB (2001) Respiratory syncytial virus and parainfluenza virus. New England Journal of Medicine 344, 1917–1928. [DOI] [PubMed] [Google Scholar]
- 10.Murphy BR (1988) Current approaches to the development of vaccines effective against parainfluenza viruses. Bulletin of World Health Organization 66, 391–397. [PMC free article] [PubMed] [Google Scholar]
- 11.Hayden FG (2006) Respiratory viral threats. Current Opinion in Infectious Diseases 19, 169–178. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg GA et al. (2009) Parainfluenza virus infection of young children: estimates of the population-based burden of hospitalization. Journal of Pediatrics 154, 694–699. [DOI] [PubMed] [Google Scholar]
- 13.Rudan I et al. (2010) Causes of deaths in children younger than 5 years in China in 2008. Lancet 375, 1083–1089. [DOI] [PubMed] [Google Scholar]
- 14.Chanock RM (1956) Association of a new type of cytopathogenic myxovirus with infantile croup. Journal of Experimental Medicine 104, 555–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laurichesse H et al. (1999) Epidemiological features of parainfluenza virus infections: laboratory surveillance in England and Wales, 1975–1997. European Journal of Epidemiology 15, 475–484. [DOI] [PubMed] [Google Scholar]
- 16.Reed G et al. (1997) Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children 5 years old. Journal of Infectious Diseases 175, 807–813. [DOI] [PubMed] [Google Scholar]
- 17.Yano T et al. (2014) Epidemiological investigation and seroprevalence of human parainfluenza virus in Mie Prefecture in Japan during 2009–2013. Japanese Journal of Infectious Diseases 67, 506–508. [PubMed] [Google Scholar]
- 18.Moisiuk SE et al. (1998) Outbreak of parainfluenza virus type 3 in an intermediate care neonatal nursery. Pediatric Infectious Diseases Journal 17, 49–53. [DOI] [PubMed] [Google Scholar]
- 19.Cortez KJ et al. (2001) Outbreak of human parainfluenza virus 3 infections in a hematopoietic stem cell transplant population. Journal of Infectious Diseases 184, 1093–1097. [DOI] [PubMed] [Google Scholar]
- 20.Cohen BJ et al. (2007) Plaque reduction neutralization test for measles antibodies: description of a standardised laboratory method for use in immunogenicity studies of aerosol vaccination. Vaccine 26, 59–66. [DOI] [PubMed] [Google Scholar]
- 21.Arnold K et al. (2006) The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics (oxford, England) 22, 195–201. [DOI] [PubMed] [Google Scholar]
- 22.Bordoli L et al. (2009) Protein structure homology modelling using SWISS-MODEL workspace. Nature Protocols 4, 1–13. [DOI] [PubMed] [Google Scholar]
- 23.Biasini M et al. (2014) SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Research 42, 252–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kringelum JV et al. (2012) Reliable B cell epitope predictions: impacts of method development and improved benchmarking. PLoS Computational Biology 8, e1002829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ponomarenko J et al. (2008) Ellipro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinformatics 9, 514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jespersen MC, et al. (2017) BepiPred-2.0: improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Research 45, W24–W29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamby SE and Hirst JD (2008) Prediction of glycosylation sites using random forests. BMC Bioinformatics 9, 500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delport W et al. (2010) Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics (oxford, England) 26, 2455–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Numazaki Y et al. (1968) A variant of parainfluenza type 2 virus. Proceedings of the Society for Experimental Biology and Medicine 127, 992–996. [DOI] [PubMed] [Google Scholar]
- 30.Tsurudome M et al. (1989) Extensive antigenic diversity among human parainfluenza type 2 virus isolates and immunological relationships among paramyxoviruses revealed by monoclonal antibodies. Virology 171, 38–48. [DOI] [PubMed] [Google Scholar]
- 31.Ray R, et al. (1992) Distinct hemagglutinin and neuraminidase epitopes involved in antigenic variation of recent human parainfluenza virus type 2 isolates. Virus Research 24, 107–113. [DOI] [PubMed] [Google Scholar]
- 32.Santibanez S et al. (2002) Rapid replacement of endemic measles virus genotypes. Journal of General Virology 83, 2699–2708. [DOI] [PubMed] [Google Scholar]
- 33.Chen YY et al. (2011) Japanese encephalitis virus genotype replacement, Taiwan, 2009–2010. Emerging Infectious Diseases 17, 2354–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Do LP, et al. (2015) Molecular epidemiology of Japanese encephalitis in northern Vietnam, 1964–2011: genotype replacement. Virology Journal 12, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spriggs MK et al. (1987) Expression of the F and HN glycoproteins of human parainfluenza virus type 3 by recombinant vaccinia viruses: contributions of the individual proteins to host immunity. Journal of Virology 61, 3416–3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Örvell C et al. (1997) Characterization of genotype-specific epitopes of the HN protein of mumps virus. Journal of General Virology 78, 3187–3193. [DOI] [PubMed] [Google Scholar]
- 37.Cusi MG et al. (2001) Localization of a new neutralizing epitope on the mumps virus hemagglutinin-neuraminidase protein. Virus Research 74, 133–137. [DOI] [PubMed] [Google Scholar]
- 38.Bouche FB, Ertl OT and Muller CP (2002) Neutralizing B cell response in measles. Viral Immunology 15, 451–471. [DOI] [PubMed] [Google Scholar]
- 39.Samal SK (2006) Paramyxoviruses of animals. In Mahy BWJ and van Regenmortel MHV (eds), Desk Encyclopedia of Animal and Bacterial Virology. Oxford: Elsevier, pp. 175–182. [Google Scholar]
- 40.Šantak M, Örvell C and Gulija TK (2015) Identification of conformational neutralization sites on the fusion protein of mumps virus. Journal of General Virology 96, 982–990. [DOI] [PubMed] [Google Scholar]
- 41.Schmidt AC et al. (2011) Progress in the development of human parainfluenza virus vaccines. Expert Review in Respiratory Medicine 5, 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268818001693.
click here to view supplementary material