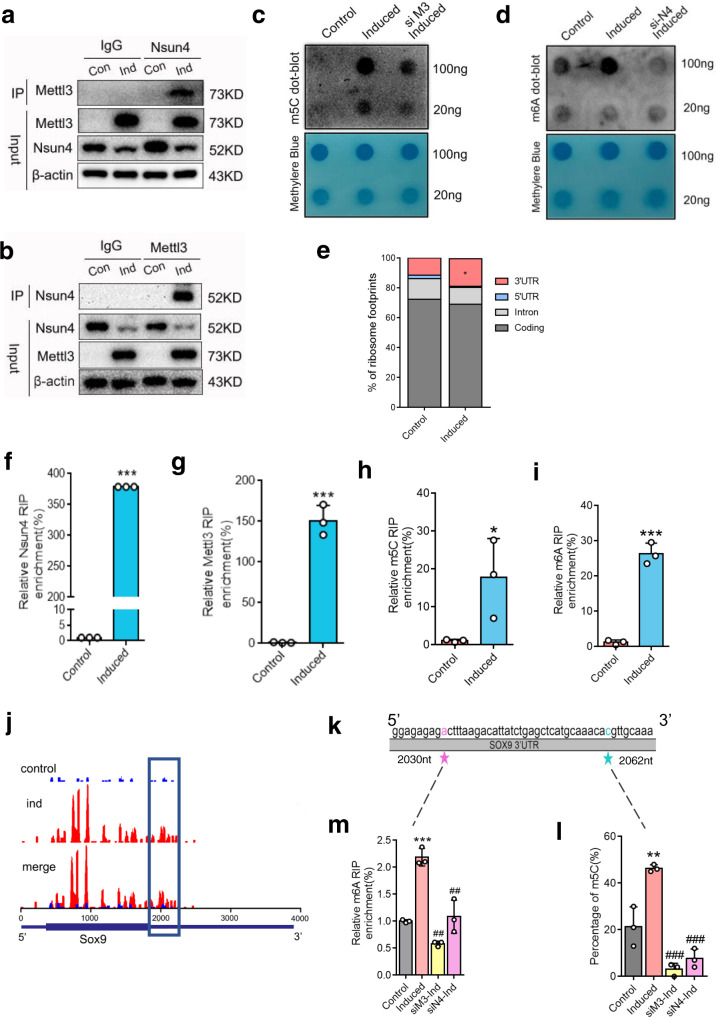
Fig. 3. Nsun4 and Mettl3 co-regulate m5C and m6A methylation of Sox9 depends by binding to its 3’UTR.
a Proteins were immunoprecipitated with Nsun4 antibody from BMSCs with or without undergoing chondrogenic differentiation. Mettl3 was detected with the indicated antibody. b Proteins were immunoprecipitated with Mettl3 antibody from BMSCs with or without undergoing chondrogenic differentiation; Nsun4 was detected with the indicated antibody. c, d m5C c and m6A d dot blot of the Mettl3 or Nsun4 knockdown BMSCs undergoing chondrogenic differentiation. e Proportion of ribosome footprints distribution in the 5’UTR, 3’UTR, Coding or Intron region across the entire set of mRNA transcripts. f, g Binding of Nsun4, Mettl3 with the 3’UTR of Sox9 mRNA in control and induced cells were analyzed by Nsun4 (f) or Mettl3 (g) RIP-qPCR. h, i m5C (h) and m6A (i) RIP-qPCR analysis of the 3’UTR of Sox9 mRNA in control or induced cells. j Ribosomes were enriched in 3’UTRs of Sox9 genes from Ribo-seq data. Square marked increases of Ribosome Footprints in chondroblast cells undergoing chondrogenic differentiation. n = 3. k Schematic depiction of the m5C and m6A sites. l Statistical analysis for the percentage of the m5C site in the 3’UTR of the Sox9 mRNA by RNA-BisSeq. m BMSCs were transfected with or without siRNAs for 24 h, then cultured in chondrogenic induce medium; the m6A site in Sox9 3’UTR was analyzed by m6A RIP-qPCR. Data are presented as means ± SD from three independent experiment. Student’s t test were performed for (e–i). One way ANOVA and Tukey’s multiple comparison tests were performed for (m, l). *P < 0.05, **p < 0.01, ***p < 0.001 compared with the control group; ##p < 0.01, ###p < 0.001 compared with the induced group.