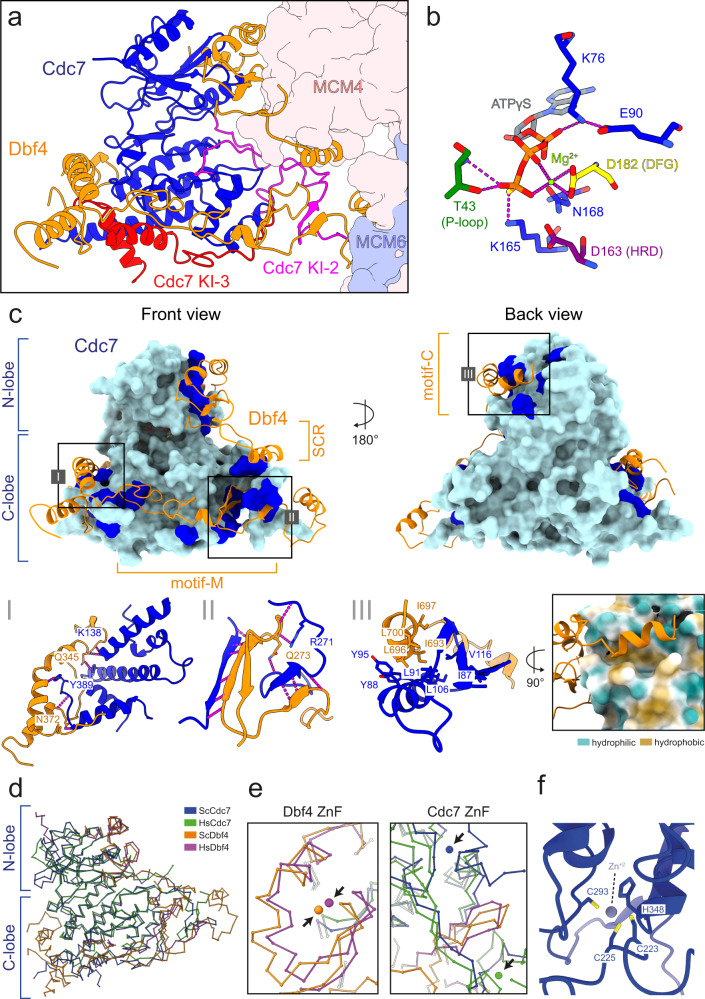
Fig. 3. DDK organization and intra-molecular interactions.
a Front view of DDK bound to the DH. The Cdc7 kinase insert (KI) regions are indicated, KI-2 in magenta and KI-3 in red. b Zoomed view of the Cdc7 active site bound to ATPγS and a Mg2+ ion. The Cdc7 residues surrounding the nucleotide are shown. D182 (DFG motif), T43 (P-loop), and D163 (HRD motif) are coloured in yellow, dark green and purple, respectively and other Cdc7 residues are coloured in blue. c Front and back view of the core of DDK. The surface of Cdc7 is coloured in cyan and regions which contact Dbf4 are highlighted in blue. Zoomed view of the interaction interfaces between: I Dbf4 motif-M and a helical bundle within the Cdc7 C-lobe, II Dbf4 motif-M and Cdc7 KI-3 and III Dbf4 motif-C and a hydrophobic groove on the surface of the Cdc7 N-lobe. a–c Cdc7 and Dbf4 are coloured in blue and orange, respectively and hydrogen bond interactions are represented by dotted magenta-coloured lines. d Overall 3D organization of the core of human DDK (PDB:6YA7) and budding yeast DDK (MD-(ATPγS)). Both kinase structures are highly similar and display an active kinase conformation. e Overlay of the zinc finger (ZnF) domains of Dbf4 motif-C and Cdc7 from human and budding yeast. The Dbf4 ZnF is at same 3D position and the Cdc7 ZnF is located at the back of Cdc7 but deviates by 21 Å. f Zoomed view of the budding yeast Cdc7 ZnF and the residues involved in zinc ion coordination.