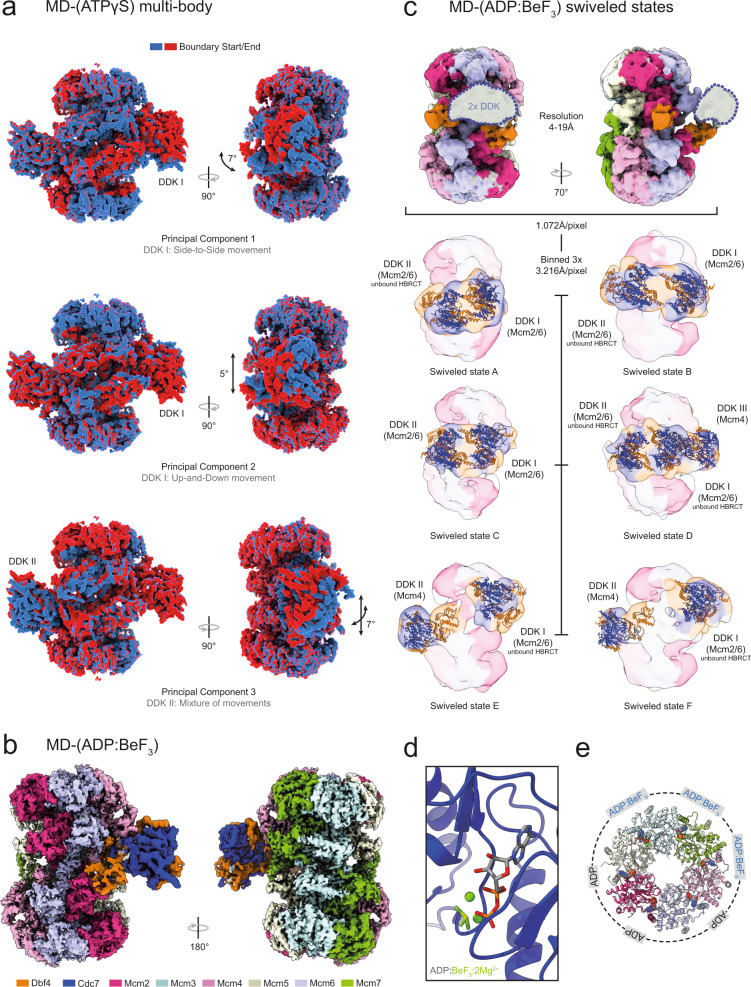
Fig. 6. DDK dynamics revealed through multi-body refinement and flexible analysis and alternative MD complex conformations in the presence of ADP:BeF3.
a Flexible analysis of the multi-body refinement of MD-(ATPγS) state III shows distinct movements of DDK relative to the MCM2-7 DH. In addition to DDK movement, the DH C-terminal domain also shows rotational movements. b Side and top views of the cryo-EM composite map (see Methods) of the MD complex in the presence of ADP:BeF3. DH at 3.8 Å mean resolution and DDK at 4.4 Å mean resolution. c The alternative swiveled structural states of the MD complex in the presence of ADP:BeF3. The EM map derived from the unbinned data displayed density large enough to fit two DDK subunits in a swiveled conformation, but the data had to be Fourier binned 3 × 3 to obtain easier to interpret EM maps. The data revealed a range of DDK binding modes, that feature in some cases either or both Mcm2/6 and Mcm4 targeted DDK conformations. The Mcm subunit which is targeted by each DDK is labelled. In some states, the HBRCT domain of Dbf4 is not bound to Mcm2. The local resolution EM maps of the different MD-(ADP:BeF3) swiveled states are shown, coloured according to the key shown in (b). The atomic model of DDK/Dbf4-HBRCT, derived from the map shown in (b), was manually docked into the EM maps. d Zoomed view of the Cdc7 active site bound to ADP:BeF3 and 2 Mg2+ ions. e Overview of the nucleotide occupancy and type in each Mcm subunit within the MD-(ADP:BeF3) complex.