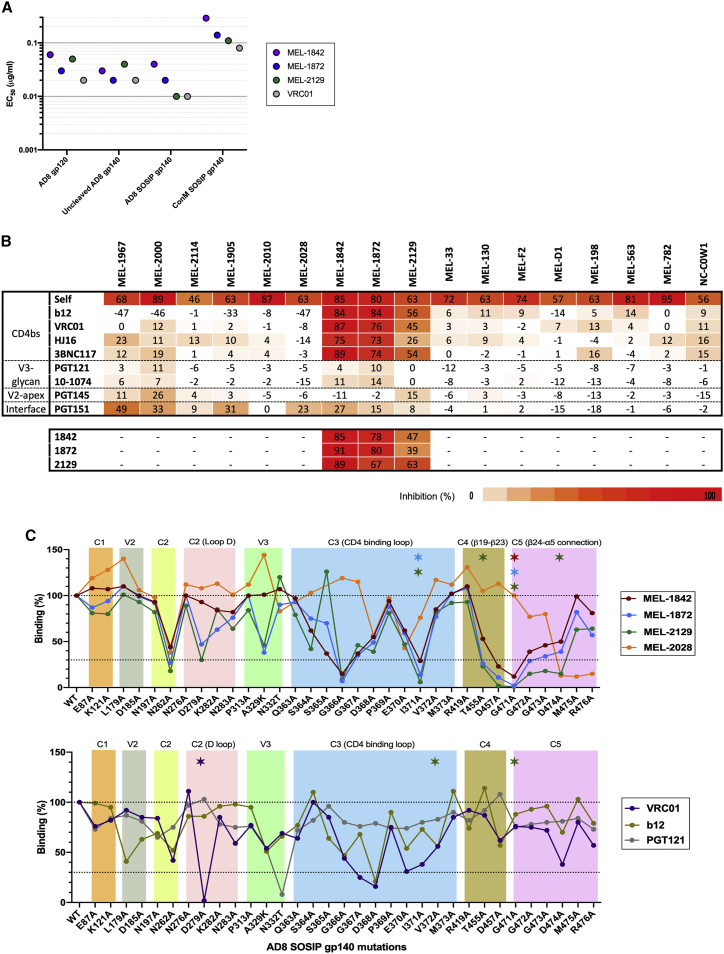
Figure 5.
Epitope mapping of bovine monoclonal antibodies
(A) Binding of bovine bNAbs to HIV Envs. Bovine bNAbs were tested in direct ELISA assays to evaluate their binding to different forms of Env (monomeric gp120, uncleaved gp140, and SOSIP gp140) as well as ConM SOSIP, which is an Env trimer based on a consensus sequence of all HIV-1 group M isolates.
(B) Competition of bovine bNAbs with human bNAb for Env binding. The table shows the competition ELISA assay with values demonstrating Env binding inhibition (percentage) of human bNAbs by bovine antibodies. Competition ELISA between antibodies MEL-1842, MEL-1872, and MEL-2129 showed these antibodies bind to the same or proximate epitopes. Higher competition is shown in red, and lower inhibition values are in increasingly pale shades of orange. ELISA assays were performed in duplicates with two independent biological replicates.
(C) The effect of alanine mutagenesis on binding of bovine antibodies to AD8 gp120 captured from lysed virions. ELISA assay was performed using a constant half-maximal effective concentration (EC50) of each antibody to AD8 WT Env. Stars also show significant neutralization IC50 increase of each antibody against mutated virus compared with WT virus (5-fold for all mAbs, except 198). PGT121 (V3-glycan epitope) and b12 and VRC01 (CD4bs epitope) were included for comparison. ELISA and neutralization assays were performed in duplicate with two independent biological replicates.