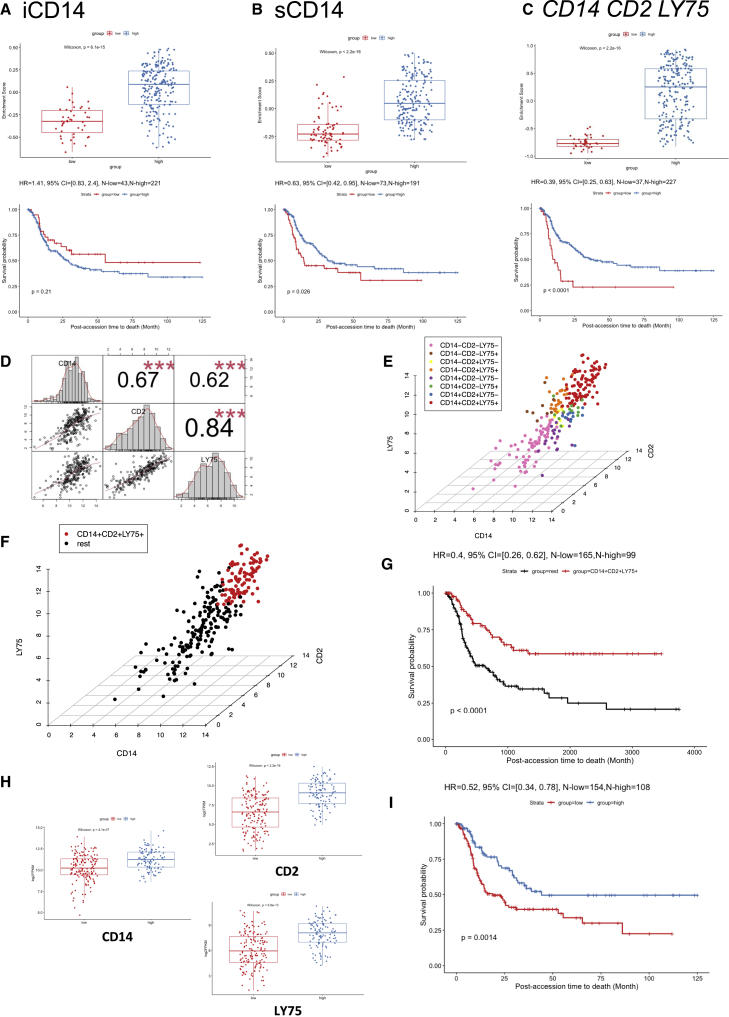
Figure 5.
Survival analysis in TCGA cohorts
(A) Survival analysis of DEGs up-regulated in iCD14+ cells in curated TCGA metastatic melanoma cohort. The upper panel shows the boxplot of the gene set enrichment score of the two patient groups stratified by the expression level of DEGs up-regulated in iCD14+ cells (see details in STAR Methods). The patient group with higher and lower median gene set enrichment score is named as the “high” and “low” group, respectively. Nonparametric test p value is indicated in the boxplot. The lower panel shows the long-term survival curve for the two groups of patients in the TCGA metastatic melanoma cohort. The two groups do not have significantly different survival outcome, with p value = 0.21. The hazard ratio value, 95% confidence interval, and number of patients in the “high” and “low” groups are indicated in the plot.
(B) Similar plot as (A), but for DEGs up-regulated in sCD14+ cells. The long-term survival curve for the two groups of patients in the TCGA metastatic melanoma cohort show significantly different survival outcome, with p value = 0.026 and hazard ratio = 0.63, where the group with “high” sCD14 gene set enrichment score has better survival outcome than the group with “low” sCD14 gene set enrichment score.
(C) Similar plot as (A) but for three genes: CD14, CD2, and LY75. The long-term survival curve for the two groups of patients in the TCGA metastatic melanoma cohort show significantly different survival outcome, with p value < 0.0001 and hazard ratio = 0.39, where the group with “high” gene set enrichment score of CD14, CD2, and LY75 has better survival outcome than the group with “low” gene set enrichment score.
(D) Correlation analysis of CD14, CD2, and LY75 in the TCGA metastatic melanoma cohort. The gene expression of CD14, CD2, and LY75 are in log2 FPKM values. The histograms of the expression distribution of each of the three genes, the pairwise scatterplots of any two of the three genes, and the Pearson’s correlation coefficient with p value of any two of the three genes are shown in the plot. Three red asterisks indicate the p value is less than 0.001.
(E) 3D scatterplot of gene expression of CD14, CD2, and LY75 in the TCGA metastatic melanoma cohort. The x axis, y axis, and z axis indicate the log2 FPKM of CD14, CD2, and LY75, respectively. The mean expression value of CD14, CD2, and LY75 are used as the thresholds to define eight groups of patients (CD14+CD2+LY75+, CD14+CD2+LY75−, CD14+CD2−LY75+, CD14+CD2−LY75−, CD14−CD2+LY75+, CD14−CD2+LY75−, CD14−CD2−LY75+, and CD14−CD2−LY75−). The colors of the points indicate the group that a patient belongs to.
(F) Similar plot as (E) but for CD14+CD2+LY75+ versus the rest of patients in the TCGA metastatic melanoma cohort. The CD14+CD2+LY75+ and the rest of the patients are highlighted by red and black, respectively.
(G) The long-term survival curve for the two groups of patients defined in (F). The two groups have significantly different survival outcome with p value <0.0001. The hazard ratio value, 95% confidence interval, and number of patients in the CD14+CD2+LY75+ and “rest” groups are indicated in the plot.
(H) Expression comparisons between low lymphocyte density group and high lymphocyte density group for CD14, CD2, and LY75, respectively. Nonparametric test p value is indicated in each plot.
(I) The survival analysis between the low lymphocyte density group and the high lymphocyte density group, where the high group has better survival outcome than the low group with HR = 0.52 and p value = 0.0014.