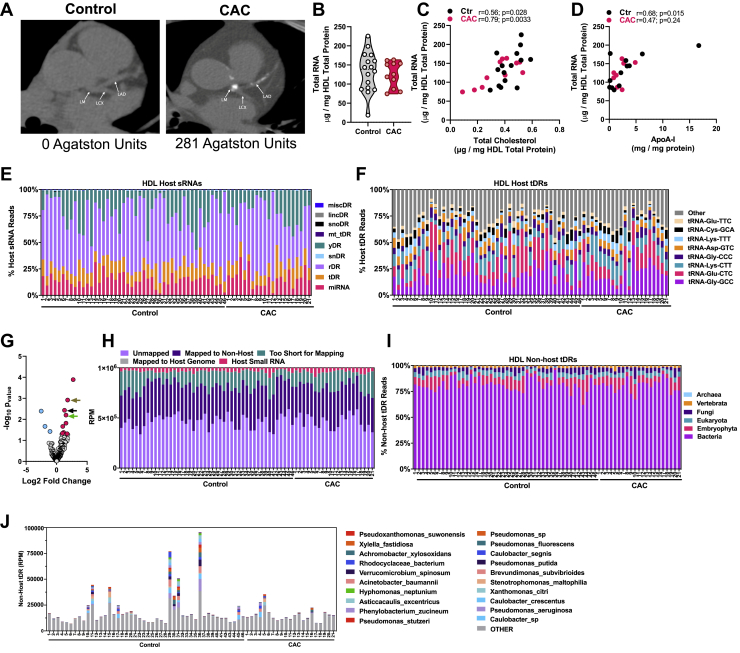
Figure 1.
Distribution of small noncoding RNAs on human HDL.A, noncontrast CT (computed tomography) scans from representative control and individual with calcified coronary atheroma and CAC score >0. Left, CAC=0; right, CAC=281. Arrows indicate LAD (left anterior descending coronary artery), LCX (left circumflex artery), and LM (left main coronary artery). B, total RNA content as determined by direct SYTO RNASelect staining (50 μM) on DGUC-HDL from control (n = 16) and CAC subjects (n = 11), normalized to total protein. Violin plot showing median and quartile ranges. C, correlation of total RNA content from (B) and total cholesterol levels normalized to total protein. D, correlation of Total RNA content (B) and apoA-I protein (ELISA) normalized to total protein. Results from sRNA-seq analysis of DGUC-HDL in control (n = 46) and CAC (n = 21) subjects. E, distribution of host sRNAs abundance as percentage of total host reads. F, percentage of host tDR by anticodon. G, differentially altered tDRs reads. Altered tDR-GlyGCC reads indicated with arrows, brown=tDR-GlyGCC-30; black=tDR-GlyGCC-33; green=tDR-GlyGCC-32. H, reads per million (RPM) identified as host, nonhost, genome, unmapped, or too short for conservative mapping with TIGER (7). I, alignment to nonhost (nonhuman) tDR database grouped by taxa. J, alignment to top nonhost tDR bacteria species. CAC, coronary artery calcium; DGUC, density-gradient ultracentrifugation; HDL, high-density lipoprotein; tDR, tRNA-derived sRNA; sRNAs, small RNAs.