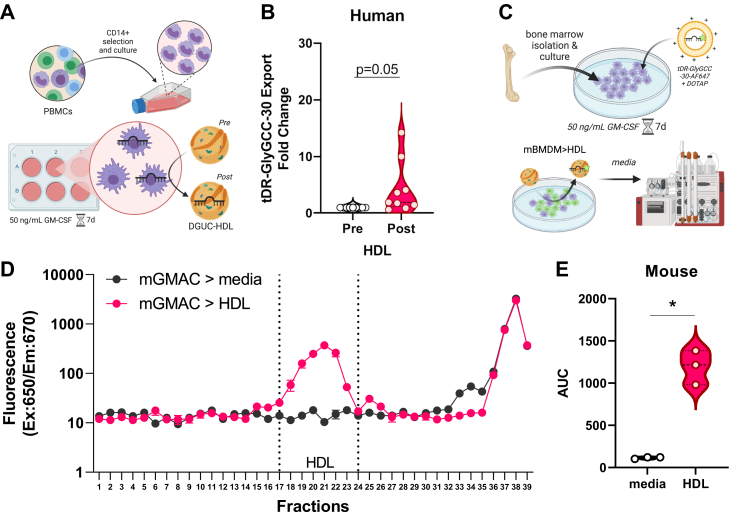
Figure 2.
Macrophage export of sRNAs to HDL.A, schematic representation of experimental design used in (B). B, HDL tDR-GlyGCC-30 expression by real-time PCR before (Pre) and after (Post) exposure to human GM-CSF differentiated macrophages. Violin plot showing median and quartile ranges, n = 9. Mann–Whitney nonparametric test. ∗p < 0.05. C, schematic representation of experimental design used in (D–E). D, fluorescence signal of exported tDR-GlyGCC-30 (AF647) from mouse bone marrow–derived macrophages (mGMAC) to serum free media (mGMAC>media) or DGUC-HDL (mGMAC>HDL) across SEC fractions (1.5 ml), data are mean ± SD, n = 3. E, area under the curve (AUC) of HDL region from (D). Violin plot showing median and quartile ranges. Unpaired t test. ∗p < 0.05. Schematics created with BioRender.com. DGUC, density-gradient ultracentrifugation; HDL, high-density lipoprotein; tDR, tRNA-derived sRNA; sRNAs, small RNAs.